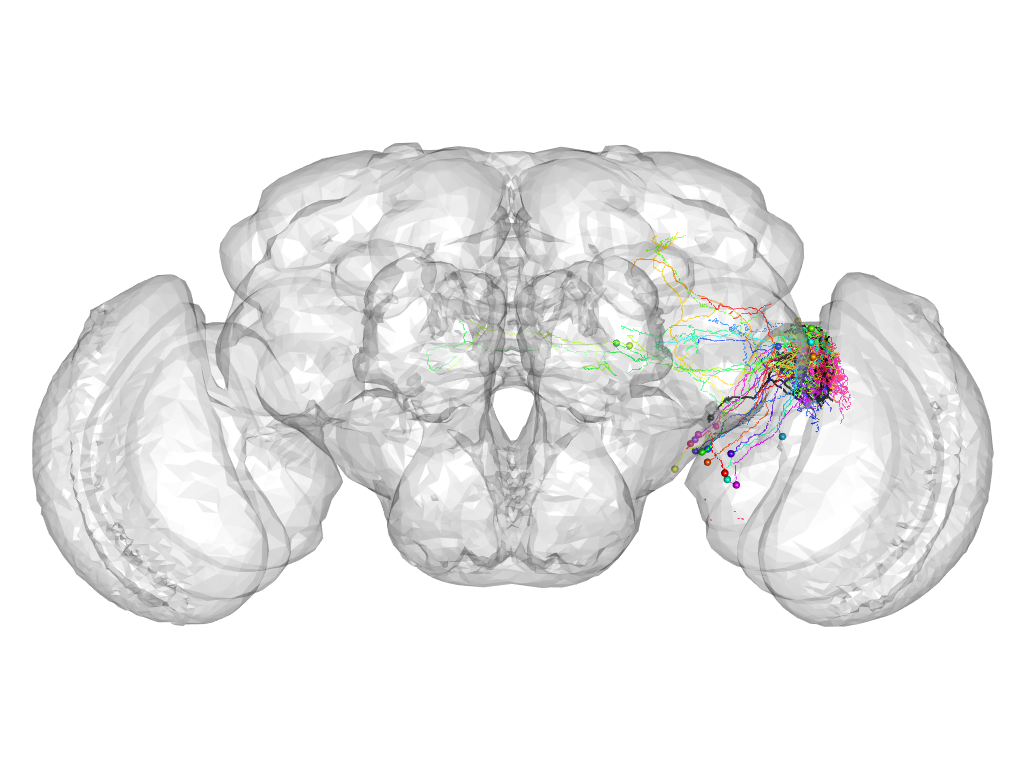
Cluster 256
Part of supercluster I
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Cha-F-000296), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 775 | VGlut-F-500422 | II | 0.78 |
| 13 | fru-M-500232 | I | 0.71 |
| 451 | VGlut-F-300582 | I | 0.58 |
| 509 | VGlut-F-200536 | VII | 0.55 |
| 327 | Cha-F-100171 | I | 0.55 |
| 406 | fru-F-000053 | VII | 0.52 |
| 293 | fru-F-000074 | II | 0.51 |
| 490 | Cha-F-000064 | I | 0.51 |
Cluster composition
This cluster has 23 neurons. The exemplar of this cluster (Cha-F-000296) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Cha-F-000296 | 4186 | ChaMARCM-F001290_seg002 | Cha-Gal4 | F | 53506.60 | 1.00 |
| fru-M-400208 | 127 | FruMARCM-M002323_seg001 | fru-Gal4 | M | 25627.71 | 0.46 |
| Cha-F-100355 | 3883 | ChaMARCM-F001547_seg001 | Cha-Gal4 | F | 33224.69 | 0.64 |
| Gad1-F-000097 | 4109 | GadMARCM-F000694_seg001 | Gad1-Gal4 | F | 35042.61 | 0.60 |
| Cha-F-000237 | 5088 | ChaMARCM-F001150_seg001 | Cha-Gal4 | F | 31438.70 | 0.45 |
| Cha-F-400163 | 5347 | ChaMARCM-F000719_seg001 | Cha-Gal4 | F | 27030.63 | 0.54 |
| Cha-F-400205 | 5661 | ChaMARCM-F000930_seg001 | Cha-Gal4 | F | 34852.66 | 0.48 |
| Gad1-F-200051 | 6009 | GadMARCM-F000194_seg001 | Gad1-Gal4 | F | 37276.49 | 0.57 |
| Gad1-F-000055 | 6151 | GadMARCM-F000507_seg001 | Gad1-Gal4 | F | 33048.77 | 0.68 |
| Cha-F-700105 | 7576 | ChaMARCM-F000435_seg001 | Cha-Gal4 | F | 31101.41 | 0.40 |
| Cha-F-300032 | 8059 | ChaMARCM-F000131_seg001 | Cha-Gal4 | F | 33125.49 | 0.48 |
| VGlut-F-300502 | 8768 | DvGlutMARCM-F003588_seg001 | VGlut-Gal4 | F | 28221.08 | 0.54 |
| VGlut-F-600663 | 9005 | DvGlutMARCM-F003549_seg001 | VGlut-Gal4 | F | 36062.64 | 0.68 |
| VGlut-F-500670 | 9088 | DvGlutMARCM-F003404_seg002 | VGlut-Gal4 | F | 20196.91 | 0.52 |
| Gad1-F-400107 | 9718 | GadMARCM-F000561_seg001 | Gad1-Gal4 | F | 29746.59 | 0.48 |
| Gad1-F-600007 | 10581 | GadMARCM-F000099_seg002 | Gad1-Gal4 | F | 28331.16 | 0.57 |
| Gad1-F-300051 | 10700 | GadMARCM-F000199_seg001 | Gad1-Gal4 | F | 25094.71 | 0.58 |
| VGlut-F-700242 | 11186 | DvGlutMARCM-F003034_seg001 | VGlut-Gal4 | F | 23917.45 | 0.55 |
| VGlut-F-400475 | 12941 | DvGlutMARCM-F1976_seg2 | VGlut-Gal4 | F | 20761.89 | 0.53 |
| VGlut-F-300314 | 15304 | DvGlutMARCM-F1638_seg2 | VGlut-Gal4 | F | 26986.37 | 0.52 |
| VGlut-F-300331 | 15390 | DvGlutMARCM-F1748_seg1 | VGlut-Gal4 | F | 25796.76 | 0.57 |
| VGlut-F-200147 | 15563 | DvGlutMARCM-F696-X2_seg1 | VGlut-Gal4 | F | 25780.51 | 0.55 |
| VGlut-F-400046 | 15876 | DvGlutMARCM-F279-X2_seg1 | VGlut-Gal4 | F | 29375.28 | 0.62 |