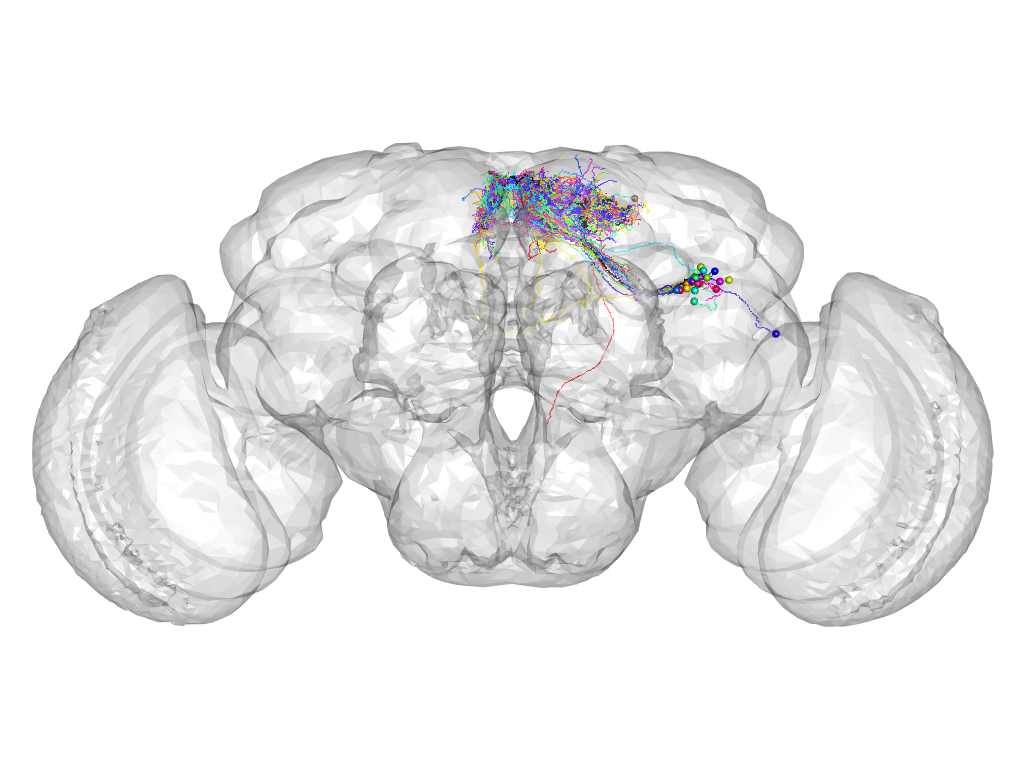
Cluster 153
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-000130), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 886 | Trh-F-300042 | XIX | 0.69 |
| 199 | TH-M-000044 | XIX | 0.68 |
| 206 | Trh-M-000018 | XIX | 0.62 |
| 856 | TH-F-200079 | XVIII | 0.57 |
| 663 | Tdc2-F-200026 | XIX | 0.54 |
| 282 | Cha-F-100339 | XVII | 0.53 |
| 40 | fru-M-000123 | XIX | 0.51 |
| 461 | Cha-F-900029 | XVII | 0.51 |
| 880 | Trh-F-500020 | XIX | 0.50 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (Trh-M-000130) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-000130 | 2316 | TPHMARCM-M001798_seg001 | Trh-Gal4 | M | 27710.00 | 1.00 |
| fru-M-600081 | 629 | FruMARCM-M001840_seg001 | fru-Gal4 | M | 7327.01 | 0.23 |
| Trh-M-000142 | 1734 | TPHMARCM-M001811_seg001 | Trh-Gal4 | M | 19933.69 | 0.71 |
| Trh-M-100041 | 1982 | TPHMARCM-0490M_seg001 | Trh-Gal4 | M | 20282.35 | 0.72 |
| Trh-M-900058 | 2192 | TPHMARCM-M001682_seg002 | Trh-Gal4 | M | 20743.50 | 0.73 |
| Trh-M-000096 | 2284 | TPHMARCM-M001765_seg001 | Trh-Gal4 | M | 19035.94 | 0.62 |
| Trh-M-000101 | 2288 | TPHMARCM-M001770_seg001 | Trh-Gal4 | M | 20373.75 | 0.71 |
| Trh-M-100078 | 2639 | TPHMARCM-M001408_seg001 | Trh-Gal4 | M | 20710.46 | 0.73 |
| Trh-M-000034 | 2672 | TPHMARCM-M001438_seg001 | Trh-Gal4 | M | 21063.62 | 0.76 |
| Trh-M-000049 | 2714 | TPHMARCM-M001478_seg001 | Trh-Gal4 | M | 20070.41 | 0.72 |
| Trh-M-100030 | 3470 | TPHMARCM-340M_seg1 | Trh-Gal4 | M | 16052.08 | 0.66 |
| Trh-M-100034 | 3474 | TPHMARCM-344M_seg1 | Trh-Gal4 | M | 16535.27 | 0.66 |
| Trh-M-000015 | 3478 | TPHMARCM-349M_seg1 | Trh-Gal4 | M | 19813.42 | 0.69 |
| Trh-F-000089 | 10067 | TPHMARCM-F001848_seg001 | Trh-Gal4 | F | 20716.78 | 0.71 |
| Trh-F-000014 | 10443 | TPHMARCM-058F_seg1 | Trh-Gal4 | F | 19442.61 | 0.70 |
| Trh-F-000044 | 12516 | TPHMARCM-1298F_seg1 | Trh-Gal4 | F | 19006.22 | 0.69 |
| Trh-F-100081 | 12534 | TPHMARCM-1314F_seg1 | Trh-Gal4 | F | 20767.93 | 0.75 |
| Trh-F-000049 | 12591 | TPHMARCM-F001387_seg001 | Trh-Gal4 | F | 20890.36 | 0.74 |
| Trh-F-100094 | 12610 | TPHMARCM-F001405_seg001 | Trh-Gal4 | F | 21419.39 | 0.76 |
| Trh-F-000004 | 13804 | TPHMARCM-022F_seg1 | Trh-Gal4 | F | 19995.01 | 0.69 |
| Trh-F-000013 | 13826 | TPHMARCM-047F_seg1 | Trh-Gal4 | F | 21177.13 | 0.74 |
| Trh-F-100034 | 13935 | TPHMARCM-224F_seg1 | Trh-Gal4 | F | 20387.79 | 0.74 |
| Trh-F-000022 | 13945 | TPHMARCM-237F_seg1 | Trh-Gal4 | F | 20517.51 | 0.73 |
| Trh-F-100036 | 14007 | TPHMARCM-295F_seg1 | Trh-Gal4 | F | 20611.93 | 0.74 |
| Trh-F-000027 | 14035 | TPHMARCM-324F_seg1 | Trh-Gal4 | F | 19943.52 | 0.72 |
| Trh-F-100041 | 14038 | TPHMARCM-328F_seg1 | Trh-Gal4 | F | 20386.05 | 0.74 |
| VGlut-F-100053 | 15455 | DvGlutMARCM-F615_seg1 | VGlut-Gal4 | F | 19405.54 | 0.70 |
| VGlut-F-000186 | 15466 | DvGlutMARCM-F626_seg1 | VGlut-Gal4 | F | 21280.74 | 0.78 |