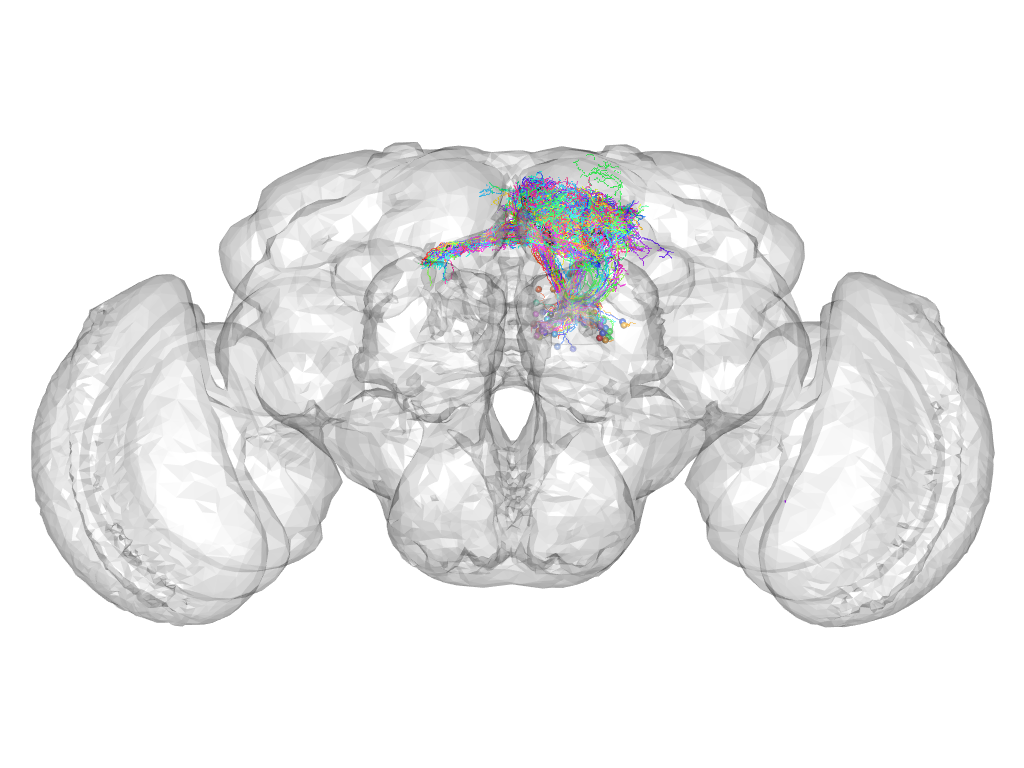
Cluster 890
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-300054), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 880 | Trh-F-500020 | XIX | 0.76 |
| 663 | Tdc2-F-200026 | XIX | 0.69 |
| 875 | Trh-F-100018 | XIX | 0.52 |
| 141 | Trh-M-400106 | XIX | 0.45 |
| 886 | Trh-F-300042 | XIX | 0.41 |
Cluster composition
This cluster has 76 neurons. The exemplar of this cluster (Trh-F-300054) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-300054 | 14081 | TPHMARCM-430F_seg1 | Trh-Gal4 | F | 12869.83 | 1.00 |
| Trh-M-600109 | 2001 | TPHMARCM-M001698_seg002 | Trh-Gal4 | M | 8869.88 | 0.70 |
| Trh-M-500188 | 2106 | TPHMARCM-M001605_seg001 | Trh-Gal4 | M | 8948.73 | 0.69 |
| Trh-M-200093 | 2128 | TPHMARCM-M001620_seg001 | Trh-Gal4 | M | 9108.93 | 0.71 |
| Trh-M-400122 | 2130 | TPHMARCM-M001621_seg001 | Trh-Gal4 | M | 9272.00 | 0.73 |
| Trh-M-400130 | 2147 | TPHMARCM-M001636_seg001 | Trh-Gal4 | M | 8496.06 | 0.67 |
| Trh-M-300153 | 2166 | TPHMARCM-M001657_seg001 | Trh-Gal4 | M | 6973.45 | 0.58 |
| Trh-M-300154 | 2170 | TPHMARCM-M001660_seg001 | Trh-Gal4 | M | 9566.37 | 0.73 |
| Trh-M-200121 | 2271 | TPHMARCM-M001758_seg001 | Trh-Gal4 | M | 8909.57 | 0.71 |
| Trh-M-300130 | 2605 | TPHMARCM-1269M_seg1 | Trh-Gal4 | M | 9179.63 | 0.70 |
| Trh-M-500123 | 2722 | TPHMARCM-M001486_seg001 | Trh-Gal4 | M | 9367.40 | 0.73 |
| Trh-M-300132 | 2724 | TPHMARCM-M001488_seg001 | Trh-Gal4 | M | 8816.11 | 0.68 |
| Trh-M-300135 | 2744 | TPHMARCM-M001510_seg001 | Trh-Gal4 | M | 9368.01 | 0.72 |
| Trh-M-300136 | 2745 | TPHMARCM-M001511_seg001 | Trh-Gal4 | M | 8205.07 | 0.68 |
| Trh-M-300110 | 2875 | TPHMARCM-1119M_seg1 | Trh-Gal4 | M | 8978.25 | 0.71 |
| Trh-M-300112 | 2885 | TPHMARCM-1127M_seg1 | Trh-Gal4 | M | 9384.59 | 0.72 |
| Trh-M-200050 | 2890 | TPHMARCM-1130M_seg1 | Trh-Gal4 | M | 9178.82 | 0.72 |
| Trh-M-300077 | 2943 | TPHMARCM-887M_seg1 | Trh-Gal4 | M | 9297.60 | 0.74 |
| Trh-M-300079 | 2945 | TPHMARCM-889M_seg1 | Trh-Gal4 | M | 8456.95 | 0.65 |
| Trh-M-300084 | 2969 | TPHMARCM-909M_seg2 | Trh-Gal4 | M | 8398.74 | 0.68 |
| Trh-M-300018 | 3515 | TPHMARCM-396M_seg1 | Trh-Gal4 | M | 9045.46 | 0.72 |
| Trh-M-300029 | 3526 | TPHMARCM-414M_seg1 | Trh-Gal4 | M | 8720.66 | 0.67 |
| Trh-M-300034 | 3531 | TPHMARCM-419M_seg1 | Trh-Gal4 | M | 9289.42 | 0.73 |
| Trh-M-100045 | 3574 | TPHMARCM-504M_seg1 | Trh-Gal4 | M | 9455.23 | 0.73 |
| Trh-M-300053 | 3590 | TPHMARCM-534M_seg1 | Trh-Gal4 | M | 8869.21 | 0.67 |
| Trh-M-300056 | 3606 | TPHMARCM-549M_seg1 | Trh-Gal4 | M | 9196.17 | 0.71 |
| Trh-M-300061 | 3635 | TPHMARCM-617M_seg1 | Trh-Gal4 | M | 8933.49 | 0.70 |
| Trh-M-300062 | 3658 | TPHMARCM-673M_seg1 | Trh-Gal4 | M | 9213.09 | 0.69 |
| Trh-F-200046 | 8397 | TPHMARCM-281F_seg1 | Trh-Gal4 | F | 9399.80 | 0.66 |
| Tdc2-F-500002 | 10298 | dTdc2MARCM-F000162_seg001 | Tdc2-Gal4 | F | 8432.41 | 0.62 |
| Tdc2-F-300020 | 10300 | dTdc2MARCM-F000163_seg001 | Tdc2-Gal4 | F | 9182.30 | 0.69 |
| Tdc2-F-300021 | 10301 | dTdc2MARCM-F000164_seg001 | Tdc2-Gal4 | F | 9674.85 | 0.70 |
| Tdc2-F-300028 | 10307 | dTdc2MARCM-F000170_seg001 | Tdc2-Gal4 | F | 9158.68 | 0.69 |
| Tdc2-F-200057 | 10327 | dTdc2MARCM-F000189_seg001 | Tdc2-Gal4 | F | 9584.50 | 0.73 |
| Tdc2-F-300030 | 10329 | dTdc2MARCM-F000191_seg001 | Tdc2-Gal4 | F | 9370.07 | 0.72 |
| Tdc2-F-300032 | 10333 | dTdc2MARCM-F000195_seg001 | Tdc2-Gal4 | F | 9901.64 | 0.74 |
| Tdc2-F-100054 | 10345 | dTdc2MARCM-F000207_seg001 | Tdc2-Gal4 | F | 9569.10 | 0.73 |
| Tdc2-F-000032 | 10358 | dTdc2MARCM-F000220_seg001 | Tdc2-Gal4 | F | 9377.28 | 0.70 |
| Tdc2-F-300043 | 10368 | dTdc2MARCM-F000229_seg002 | Tdc2-Gal4 | F | 9504.20 | 0.71 |
| Tdc2-F-100065 | 10378 | dTdc2MARCM-F000240_seg001 | Tdc2-Gal4 | F | 8748.07 | 0.66 |
| Tdc2-F-200024 | 10394 | dTdc2MARCM-F000107_seg001 | Tdc2-Gal4 | F | 8944.26 | 0.67 |
| Tdc2-F-100042 | 10414 | dTdc2MARCM-F000129_seg001 | Tdc2-Gal4 | F | 9518.91 | 0.70 |
| Tdc2-F-400012 | 10422 | dTdc2MARCM-F000138_seg001 | Tdc2-Gal4 | F | 9084.15 | 0.68 |
| Tdc2-F-400016 | 10426 | dTdc2MARCM-F000141_seg001 | Tdc2-Gal4 | F | 9067.97 | 0.69 |
| Tdc2-F-200037 | 10431 | dTdc2MARCM-F000147_seg001 | Tdc2-Gal4 | F | 9250.94 | 0.70 |
| Trh-F-300143 | 12428 | TPHMARCM-1223F_seg1 | Trh-Gal4 | F | 9458.12 | 0.73 |
| Trh-F-100076 | 12517 | TPHMARCM-1299F_seg1 | Trh-Gal4 | F | 9421.45 | 0.73 |
| Tdc2-F-200008 | 12633 | dTdc-2MARCM-F-051_seg1 | Tdc2-Gal4 | F | 9141.62 | 0.71 |
| Tdc2-F-100024 | 12636 | dTdc-2MARCM-F-055_seg1 | Tdc2-Gal4 | F | 9338.43 | 0.70 |
| Tdc2-F-300010 | 12647 | dTdc-2MARCM-F-066_seg1 | Tdc2-Gal4 | F | 8938.57 | 0.71 |
| Tdc2-F-300011 | 12648 | dTdc-2MARCM-F-067_seg1 | Tdc2-Gal4 | F | 9391.11 | 0.72 |
| Tdc2-F-200017 | 12670 | dTdc2MARCM-F000092_seg001 | Tdc2-Gal4 | F | 9422.23 | 0.72 |
| Tdc2-F-300016 | 12675 | dTdc2MARCM-F000096_seg002 | Tdc2-Gal4 | F | 9395.51 | 0.74 |
| Trh-F-200103 | 12685 | TPHMARCM-1095F_seg1 | Trh-Gal4 | F | 9151.62 | 0.72 |
| Trh-F-400090 | 12686 | TPHMARCM-1096F_seg1 | Trh-Gal4 | F | 8734.46 | 0.68 |
| Trh-F-300134 | 12701 | TPHMARCM-1150F_seg1 | Trh-Gal4 | F | 9330.92 | 0.71 |
| Trh-F-300139 | 12707 | TPHMARCM-1164F_seg1 | Trh-Gal4 | F | 8840.03 | 0.70 |
| Tdc2-F-400003 | 13191 | dTdc-2MARCM-F-022--montage_seg1 | Tdc2-Gal4 | F | 8301.43 | 0.65 |
| Trh-F-300104 | 13228 | TPHMARCM-715F_seg1 | Trh-Gal4 | F | 9476.93 | 0.71 |
| Trh-F-200001 | 13795 | TPHMARCM-013F_seg1 | Trh-Gal4 | F | 9402.74 | 0.71 |
| Trh-F-900000 | 13908 | TPHMARCM-176F_seg1 | Trh-Gal4 | F | 9126.91 | 0.68 |
| Trh-F-200012 | 13931 | TPHMARCM-220F_seg1 | Trh-Gal4 | F | 9613.55 | 0.69 |
| Trh-F-200016 | 13937 | TPHMARCM-229F_seg1 | Trh-Gal4 | F | 8952.99 | 0.68 |
| Trh-F-300020 | 13964 | TPHMARCM-253F_seg2 | Trh-Gal4 | F | 9456.53 | 0.72 |
| Trh-F-300021 | 13975 | TPHMARCM-265F_seg1 | Trh-Gal4 | F | 9400.16 | 0.72 |
| Trh-F-200041 | 13980 | TPHMARCM-270F_seg1 | Trh-Gal4 | F | 9040.12 | 0.67 |
| Trh-F-300025 | 13987 | TPHMARCM-276F_seg1 | Trh-Gal4 | F | 8916.80 | 0.67 |
| Trh-F-400042 | 14029 | TPHMARCM-314F_seg1 | Trh-Gal4 | F | 7926.18 | 0.67 |
| Trh-F-300062 | 14108 | TPHMARCM-467F_seg1 | Trh-Gal4 | F | 9128.61 | 0.72 |
| Trh-F-500041 | 14110 | TPHMARCM-468F_seg2 | Trh-Gal4 | F | 9548.34 | 0.73 |
| Trh-F-200080 | 14205 | TPHMARCM-646F_seg1 | Trh-Gal4 | F | 9333.23 | 0.71 |
| Trh-F-200083 | 14229 | TPHMARCM-681F_seg1 | Trh-Gal4 | F | 9023.92 | 0.66 |
| Trh-F-200085 | 14231 | TPHMARCM-683F_seg1 | Trh-Gal4 | F | 9095.08 | 0.71 |
| Trh-F-100059 | 14242 | TPHMARCM-695F_seg1 | Trh-Gal4 | F | 9435.92 | 0.72 |
| Trh-F-200088 | 14245 | TPHMARCM-697F_seg2 | Trh-Gal4 | F | 9605.72 | 0.73 |
| Trh-F-300099 | 14247 | TPHMARCM-698F_seg2 | Trh-Gal4 | F | 9016.15 | 0.71 |