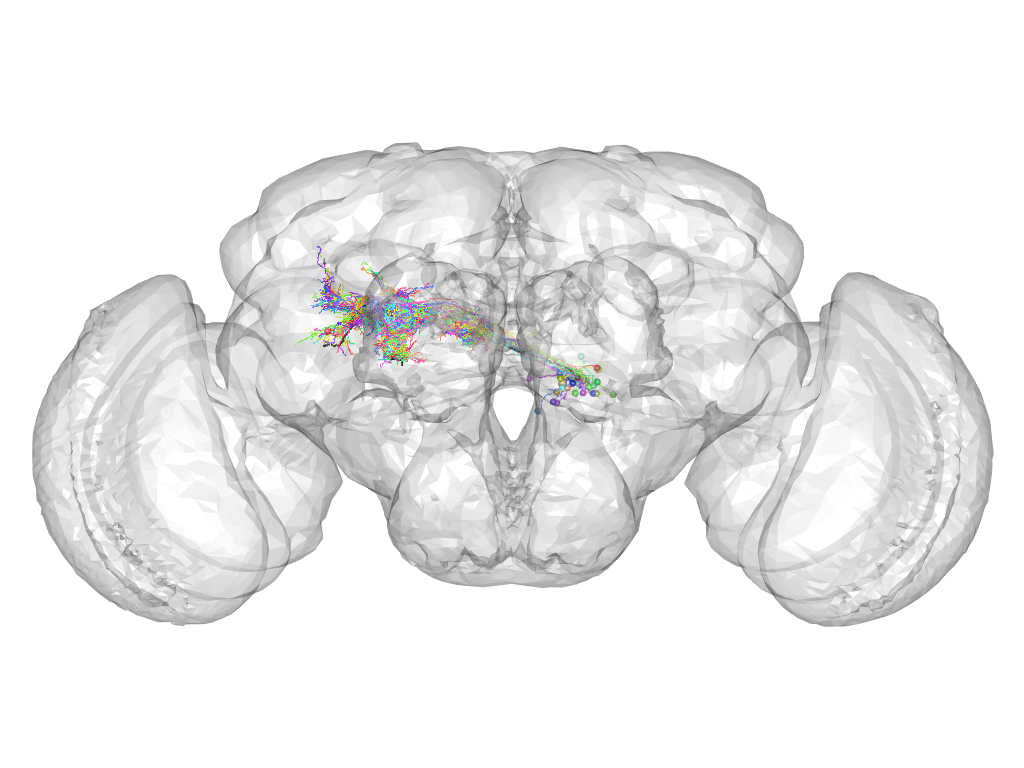
Cluster 798
Part of supercluster XIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-600079), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 166 | Trh-M-100028 | XIII | 0.67 |
| 184 | Trh-M-700031 | XIII | 0.56 |
| 82 | Trh-M-500117 | XIII | 0.53 |
| 179 | Trh-M-100061 | VI | 0.47 |
| 425 | VGlut-F-500805 | VII | 0.33 |
Cluster composition
This cluster has 41 neurons. The exemplar of this cluster (Trh-F-600079) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-600079 | 12420 | TPHMARCM-1212F_seg1 | Trh-Gal4 | F | 11981.47 | 1.00 |
| fru-M-000103 | 580 | FruMARCM-M001769_seg001 | fru-Gal4 | M | 8395.75 | 0.71 |
| Trh-M-500185 | 2090 | TPHMARCM-M001591_seg001 | Trh-Gal4 | M | 9277.91 | 0.76 |
| Trh-M-500186 | 2091 | TPHMARCM-M001591_seg002 | Trh-Gal4 | M | 8639.33 | 0.71 |
| Trh-M-500160 | 2552 | TPHMARCM-M001528_seg001 | Trh-Gal4 | M | 9219.80 | 0.74 |
| Trh-M-500173 | 2562 | TPHMARCM-M001539_seg001 | Trh-Gal4 | M | 8933.33 | 0.72 |
| Trh-M-700080 | 2616 | TPHMARCM-1326M_seg1 | Trh-Gal4 | M | 8827.33 | 0.73 |
| Trh-M-700083 | 2619 | TPHMARCM-1327M_seg2 | Trh-Gal4 | M | 8659.61 | 0.73 |
| Trh-M-500091 | 2658 | TPHMARCM-M001425_seg002 | Trh-Gal4 | M | 8763.61 | 0.73 |
| Trh-M-500068 | 2976 | TPHMARCM-916M_seg1 | Trh-Gal4 | M | 8526.45 | 0.73 |
| Trh-M-700017 | 2996 | TPHMARCM-939M_seg1 | Trh-Gal4 | M | 8789.02 | 0.73 |
| Trh-M-200006 | 3424 | TPHMARCM-118M_seg1 | Trh-Gal4 | M | 8861.99 | 0.69 |
| Trh-M-500003 | 3435 | TPHMARCM-182M_seg1 | Trh-Gal4 | M | 8568.44 | 0.69 |
| Trh-M-500023 | 3555 | TPHMARCM-486M_seg1 | Trh-Gal4 | M | 9203.38 | 0.78 |
| Trh-M-500025 | 3571 | TPHMARCM-501M_seg1 | Trh-Gal4 | M | 8813.37 | 0.74 |
| Trh-M-600020 | 3610 | TPHMARCM-562M_seg1 | Trh-Gal4 | M | 8745.48 | 0.71 |
| Trh-M-400027 | 3614 | TPHMARCM-573M_seg2 | Trh-Gal4 | M | 8854.35 | 0.75 |
| Trh-M-500044 | 3650 | TPHMARCM-664M_seg1 | Trh-Gal4 | M | 9226.44 | 0.75 |
| Trh-M-700006 | 3661 | TPHMARCM-676M_seg1 | Trh-Gal4 | M | 9016.81 | 0.75 |
| Trh-F-300078 | 5994 | TPHMARCM-592F-01_seg1 | Trh-Gal4 | F | 8815.44 | 0.75 |
| Trh-F-700052 | 12385 | TPHMARCM-1180F_seg2 | Trh-Gal4 | F | 9338.54 | 0.78 |
| Trh-F-600069 | 12395 | TPHMARCM-1193F_seg1 | Trh-Gal4 | F | 8654.41 | 0.74 |
| Trh-F-500122 | 12446 | TPHMARCM-1241F_seg1 | Trh-Gal4 | F | 8173.42 | 0.69 |
| Trh-F-500141 | 12476 | TPHMARCM-1261F_seg1 | Trh-Gal4 | F | 9002.13 | 0.74 |
| Trh-F-500142 | 12477 | TPHMARCM-1261F_seg2 | Trh-Gal4 | F | 8575.50 | 0.74 |
| Trh-F-500157 | 12501 | TPHMARCM-1285F_seg1 | Trh-Gal4 | F | 8284.29 | 0.70 |
| Trh-F-500198 | 12565 | TPHMARCM-F001354_seg001 | Trh-Gal4 | F | 8337.47 | 0.68 |
| Trh-F-400075 | 13237 | TPHMARCM-724F_seg1 | Trh-Gal4 | F | 8856.18 | 0.73 |
| Trh-F-500001 | 13848 | TPHMARCM-120F_seg1 | Trh-Gal4 | F | 9011.41 | 0.75 |
| Trh-F-500006 | 13853 | TPHMARCM-125F_seg1 | Trh-Gal4 | F | 9014.14 | 0.75 |
| Trh-F-500018 | 13866 | TPHMARCM-136F_seg1 | Trh-Gal4 | F | 8766.25 | 0.71 |
| Trh-F-200044 | 13990 | TPHMARCM-279F_seg1 | Trh-Gal4 | F | 8784.45 | 0.69 |
| Trh-F-300077 | 14148 | TPHMARCM-569F_seg1 | Trh-Gal4 | F | 8917.35 | 0.73 |
| Trh-F-600022 | 14149 | TPHMARCM-571F_seg1 | Trh-Gal4 | F | 8680.34 | 0.73 |
| Trh-F-600023 | 14150 | TPHMARCM-571F_seg2 | Trh-Gal4 | F | 8385.87 | 0.72 |
| Trh-F-600030 | 14202 | TPHMARCM-643F_seg1 | Trh-Gal4 | F | 8631.14 | 0.72 |
| Trh-F-500065 | 14217 | TPHMARCM-656F_seg1 | Trh-Gal4 | F | 9084.00 | 0.76 |
| Trh-F-600056 | 14264 | TPHMARCM-812F_seg2 | Trh-Gal4 | F | 9042.36 | 0.76 |
| Trh-F-900025 | 14274 | TPHMARCM-821F-01_seg1 | Trh-Gal4 | F | 8499.45 | 0.70 |
| Trh-F-900026 | 14275 | TPHMARCM-822F_seg1 | Trh-Gal4 | F | 9131.29 | 0.75 |
| Trh-F-900028 | 14277 | TPHMARCM-823F-01_seg1 | Trh-Gal4 | F | 8865.66 | 0.73 |