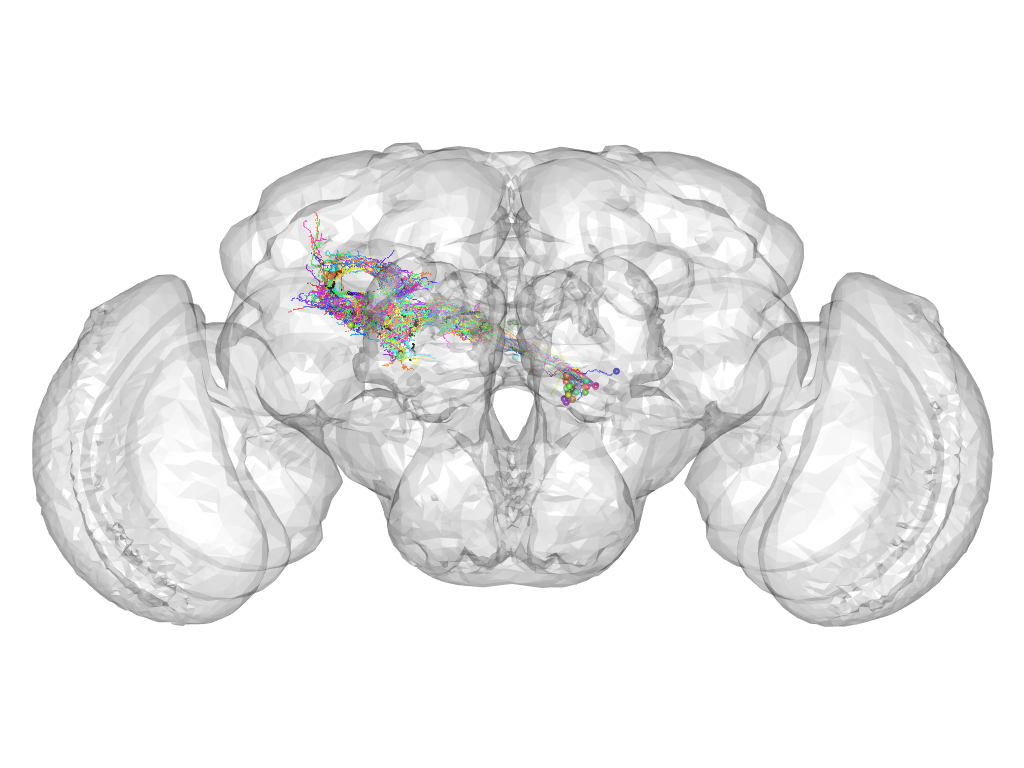
Cluster 166
Part of supercluster XIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-100028), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 184 | Trh-M-700031 | XIII | 0.71 |
| 82 | Trh-M-500117 | XIII | 0.68 |
| 798 | Trh-F-600079 | XIII | 0.63 |
| 179 | Trh-M-100061 | VI | 0.43 |
| 892 | Trh-F-300060 | VI | 0.21 |
Cluster composition
This cluster has 43 neurons. The exemplar of this cluster (Trh-M-100028) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-100028 | 2579 | TPHMARCM-108M_seg1 | Trh-Gal4 | M | 16377.71 | 1.00 |
| Trh-M-500118 | 929 | TPHMARCM-M001468_seg002 | Trh-Gal4 | M | 11207.59 | 0.70 |
| Trh-M-500166 | 1998 | TPHMARCM-M001532_seg002 | Trh-Gal4 | M | 10512.48 | 0.68 |
| Trh-M-900028 | 2035 | TPHMARCM-M001548_seg001 | Trh-Gal4 | M | 10852.87 | 0.66 |
| Trh-M-900038 | 2048 | TPHMARCM-M001554_seg002 | Trh-Gal4 | M | 11841.46 | 0.67 |
| Trh-M-600088 | 2127 | TPHMARCM-M001619_seg002 | Trh-Gal4 | M | 11653.33 | 0.73 |
| Trh-M-000100 | 2287 | TPHMARCM-M001769_seg001 | Trh-Gal4 | M | 11569.42 | 0.74 |
| Trh-M-500170 | 2559 | TPHMARCM-M001536_seg001 | Trh-Gal4 | M | 11431.73 | 0.73 |
| Trh-M-700075 | 2597 | TPHMARCM-1187M_seg1 | Trh-Gal4 | M | 10449.87 | 0.64 |
| Trh-M-600079 | 2731 | TPHMARCM-M001496_seg001 | Trh-Gal4 | M | 11756.03 | 0.73 |
| Trh-M-400078 | 2763 | TPHMARCM-1073M_seg1 | Trh-Gal4 | M | 11581.22 | 0.71 |
| Trh-M-700048 | 2794 | TPHMARCM-1023M_seg1 | Trh-Gal4 | M | 11260.42 | 0.72 |
| Trh-M-700057 | 2807 | TPHMARCM-1036M_seg1 | Trh-Gal4 | M | 11768.66 | 0.74 |
| Trh-M-900010 | 2938 | TPHMARCM-878M_seg1 | Trh-Gal4 | M | 10947.55 | 0.72 |
| Trh-M-900014 | 2948 | TPHMARCM-891M_seg2 | Trh-Gal4 | M | 11295.47 | 0.71 |
| Trh-M-900015 | 2949 | TPHMARCM-892M_seg1 | Trh-Gal4 | M | 10487.69 | 0.67 |
| Trh-M-600044 | 3001 | TPHMARCM-944M_seg1 | Trh-Gal4 | M | 11036.71 | 0.69 |
| Trh-M-600050 | 3020 | TPHMARCM-972M_seg1 | Trh-Gal4 | M | 11211.78 | 0.71 |
| Trh-M-700030 | 3038 | TPHMARCM-991M_seg1 | Trh-Gal4 | M | 11863.05 | 0.73 |
| Trh-M-400001 | 3438 | TPHMARCM-185M_seg1 | Trh-Gal4 | M | 11697.41 | 0.74 |
| Trh-M-600021 | 3611 | TPHMARCM-562M_seg2 | Trh-Gal4 | M | 11071.09 | 0.70 |
| Trh-M-900007 | 3697 | TPHMARCM-848M_seg1 | Trh-Gal4 | M | 11688.51 | 0.72 |
| Trh-F-600020 | 5992 | TPHMARCM-570F_seg1 | Trh-Gal4 | F | 11389.63 | 0.72 |
| Trh-F-600060 | 10437 | TPHMARCM-874F_seg1 | Trh-Gal4 | F | 11813.86 | 0.72 |
| Trh-F-500111 | 12406 | TPHMARCM-1201F_seg1 | Trh-Gal4 | F | 11419.48 | 0.72 |
| Trh-F-500113 | 12408 | TPHMARCM-1202F_seg1 | Trh-Gal4 | F | 11305.56 | 0.71 |
| Trh-F-600103 | 12464 | TPHMARCM-1253F_seg1 | Trh-Gal4 | F | 9465.42 | 0.66 |
| Trh-F-500143 | 12478 | TPHMARCM-1262F_seg1 | Trh-Gal4 | F | 11238.29 | 0.71 |
| Trh-F-500160 | 12504 | TPHMARCM-1286F_seg2 | Trh-Gal4 | F | 11443.04 | 0.72 |
| Trh-F-500190 | 12548 | TPHMARCM-1335F_seg1 | Trh-Gal4 | F | 11005.37 | 0.70 |
| Trh-F-500191 | 12549 | TPHMARCM-1336F_seg1 | Trh-Gal4 | F | 10876.42 | 0.69 |
| Trh-F-900032 | 12711 | TPHMARCM-854F_seg1 | Trh-Gal4 | F | 11486.65 | 0.73 |
| Trh-F-900013 | 13256 | TPHMARCM-743F_seg1 | Trh-Gal4 | F | 11584.36 | 0.72 |
| Trh-F-900024 | 13304 | TPHMARCM-786F-01_seg1 | Trh-Gal4 | F | 11451.47 | 0.72 |
| Trh-F-800005 | 13305 | TPHMARCM-787F_seg1 | Trh-Gal4 | F | 11652.71 | 0.73 |
| Trh-F-700000 | 13844 | TPHMARCM-113F_seg1 | Trh-Gal4 | F | 11107.10 | 0.71 |
| Trh-F-500005 | 13852 | TPHMARCM-124F_seg1 | Trh-Gal4 | F | 11651.16 | 0.72 |
| Trh-F-500011 | 13858 | TPHMARCM-128F_seg1 | Trh-Gal4 | F | 10906.17 | 0.68 |
| Trh-F-500012 | 13859 | TPHMARCM-129F_seg1 | Trh-Gal4 | F | 11478.37 | 0.70 |
| Trh-F-600000 | 13894 | TPHMARCM-159F_seg1 | Trh-Gal4 | F | 10934.15 | 0.68 |
| Trh-F-000021 | 13930 | TPHMARCM-219F_seg2 | Trh-Gal4 | F | 11482.44 | 0.72 |
| Trh-F-500074 | 14237 | TPHMARCM-692F_seg1 | Trh-Gal4 | F | 11488.51 | 0.73 |
| Trh-F-600058 | 14290 | TPHMARCM-833F_seg1 | Trh-Gal4 | F | 10818.19 | 0.70 |