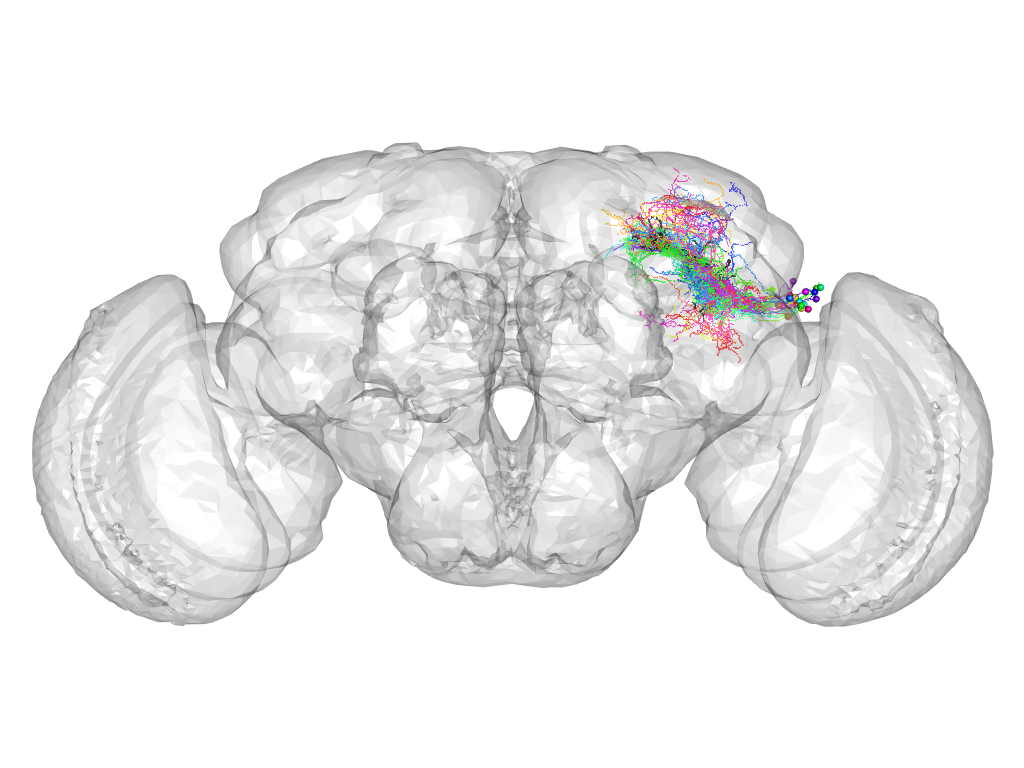
Cluster 779
Part of supercluster XVII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-800074), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 194 | TH-M-200035 | XVII | 0.68 |
| 461 | Cha-F-900029 | XVII | 0.66 |
| 268 | Cha-F-200340 | X | 0.66 |
| 282 | Cha-F-100339 | XVII | 0.63 |
| 168 | Trh-M-000025 | XVII | 0.61 |
| 621 | Gad1-F-500089 | XVII | 0.56 |
| 1006 | VGlut-F-500043 | III | 0.55 |
| 65 | fru-M-000139 | XVII | 0.55 |
| 843 | Trh-F-100060 | IX | 0.50 |
| 258 | Cha-F-100277 | XIII | 0.50 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (VGlut-F-800074) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-800074 | 12126 | DvGlutMARCM-F002621_seg002 | VGlut-Gal4 | F | 12323.15 | 1.00 |
| Cha-F-300212 | 5447 | ChaMARCM-F000786_seg002 | Cha-Gal4 | F | 5919.63 | 0.39 |
| VGlut-F-800267 | 6513 | DvGlutMARCM-F004289_seg001 | VGlut-Gal4 | F | 7298.99 | 0.57 |
| VGlut-F-700503 | 7044 | DvGlutMARCM-F004175_seg003 | VGlut-Gal4 | F | 8412.53 | 0.73 |
| VGlut-F-100348 | 7428 | DvGlutMARCM-F003882_seg001 | VGlut-Gal4 | F | 7153.50 | 0.55 |
| VGlut-F-700413 | 7957 | DvGlutMARCM-F003787_seg001 | VGlut-Gal4 | F | 7649.20 | 0.69 |
| VGlut-F-700260 | 8474 | DvGlutMARCM-F003070_seg001 | VGlut-Gal4 | F | 7172.86 | 0.51 |
| VGlut-F-500544 | 8485 | DvGlutMARCM-F003075_seg003 | VGlut-Gal4 | F | 7964.58 | 0.69 |
| VGlut-F-700280 | 8517 | DvGlutMARCM-F003098_seg002 | VGlut-Gal4 | F | 8585.31 | 0.66 |
| VGlut-F-700342 | 9107 | DvGlutMARCM-F003413_seg002 | VGlut-Gal4 | F | 7284.07 | 0.66 |
| VGlut-F-200441 | 11068 | DvGlutMARCM-F002948_seg001 | VGlut-Gal4 | F | 7500.14 | 0.59 |
| Gad1-F-400027 | 11255 | GadMARCM-F000235_seg001 | Gad1-Gal4 | F | 7269.56 | 0.60 |
| VGlut-F-600193 | 11755 | DvGlutMARCM-F002326_seg001 | VGlut-Gal4 | F | 8014.92 | 0.67 |
| VGlut-F-600218 | 11818 | DvGlutMARCM-F002381_seg001 | VGlut-Gal4 | F | 8448.58 | 0.72 |
| VGlut-F-500419 | 12010 | DvGlutMARCM-F002532_seg001 | VGlut-Gal4 | F | 7234.24 | 0.61 |
| VGlut-F-600311 | 12040 | DvGlutMARCM-F002555_seg001 | VGlut-Gal4 | F | 7998.01 | 0.65 |
| VGlut-F-800056 | 12107 | DvGlutMARCM-F002608_seg001 | VGlut-Gal4 | F | 7858.91 | 0.63 |
| VGlut-F-800057 | 12108 | DvGlutMARCM-F002608_seg002 | VGlut-Gal4 | F | 7147.82 | 0.51 |
| VGlut-F-800064 | 12115 | DvGlutMARCM-F002612_seg001 | VGlut-Gal4 | F | 7896.66 | 0.65 |
| VGlut-F-800068 | 12120 | DvGlutMARCM-F002617_seg001 | VGlut-Gal4 | F | 6037.40 | 0.47 |
| VGlut-F-500273 | 12791 | DvGlutMARCM-F1839_seg1 | VGlut-Gal4 | F | 7634.92 | 0.69 |
| VGlut-F-600122 | 13068 | DvGlutMARCM-F2091_seg1 | VGlut-Gal4 | F | 8202.58 | 0.71 |
| VGlut-F-600030 | 14482 | DvGlutMARCM-F759-x3_seg3 | VGlut-Gal4 | F | 7110.94 | 0.59 |
| VGlut-F-400240 | 14574 | DvGlutMARCM-F839_seg1 | VGlut-Gal4 | F | 8427.73 | 0.67 |
| VGlut-F-600075 | 15154 | DvGlutMARCM-F1486_seg1 | VGlut-Gal4 | F | 6492.82 | 0.46 |
| VGlut-F-600076 | 15155 | DvGlutMARCM-F1486_seg2 | VGlut-Gal4 | F | 6791.82 | 0.47 |
| VGlut-F-700001 | 15622 | DvGlutMARCM-F039_seg1 | VGlut-Gal4 | F | 7110.09 | 0.55 |
| VGlut-F-500046 | 15859 | DvGlutMARCM-F264-X2_seg1 | VGlut-Gal4 | F | 7096.60 | 0.60 |