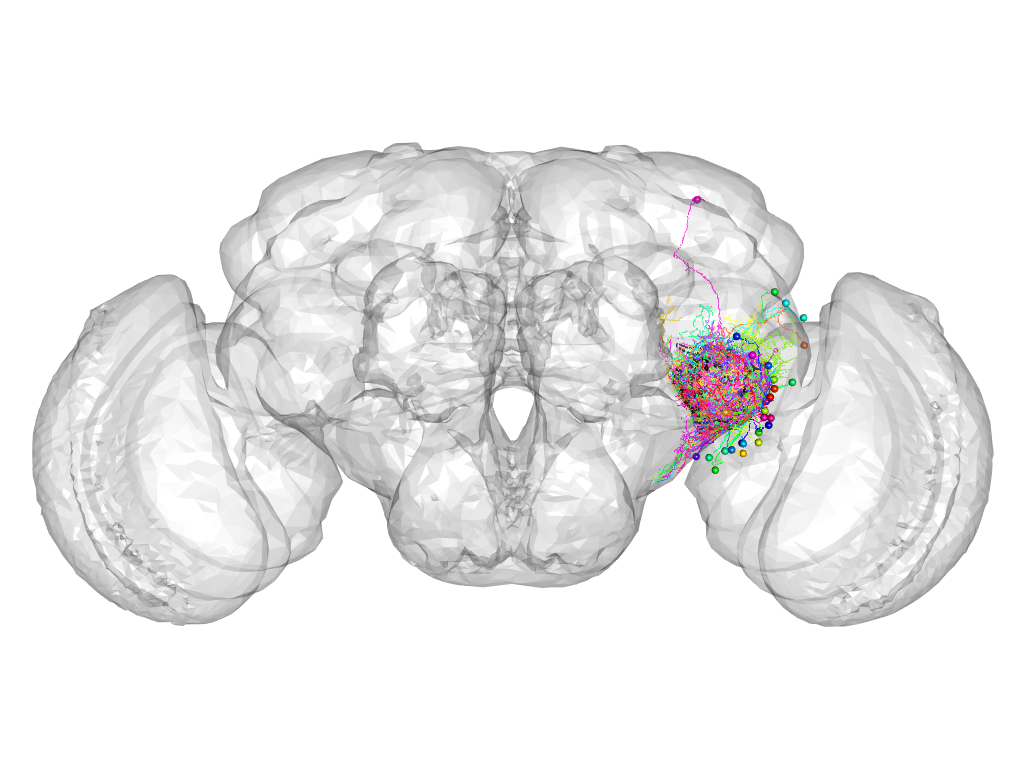
Cluster 572
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-000521), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 268 | Cha-F-200340 | X | 0.67 |
| 903 | VGlut-F-000273 | X | 0.64 |
| 876 | Trh-F-100023 | X | 0.61 |
| 250 | Gad1-F-700054 | X | 0.54 |
| 865 | TH-F-000025 | X | 0.53 |
| 200 | TH-M-000060 | X | 0.52 |
Cluster composition
This cluster has 37 neurons. The exemplar of this cluster (VGlut-F-000521) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-000521 | 8882 | DvGlutMARCM-F003463_seg001 | VGlut-Gal4 | F | 34611.87 | 1.00 |
| fru-M-700111 | 692 | FruMARCM-M001895_seg002 | fru-Gal4 | M | 13286.88 | 0.58 |
| Trh-M-600043 | 3000 | TPHMARCM-943M_seg1 | Trh-Gal4 | M | 13061.17 | 0.53 |
| Gad1-F-800070 | 4010 | GadMARCM-F000624_seg002 | Gad1-Gal4 | F | 26297.51 | 0.74 |
| Gad1-F-800080 | 4048 | GadMARCM-F000648_seg002 | Gad1-Gal4 | F | 21793.72 | 0.66 |
| Gad1-F-800085 | 4089 | GadMARCM-F000677_seg001 | Gad1-Gal4 | F | 23815.82 | 0.69 |
| Cha-F-000314 | 4245 | ChaMARCM-F001330_seg001 | Cha-Gal4 | F | 24360.28 | 0.67 |
| Cha-F-200341 | 4308 | ChaMARCM-F001372_seg001 | Cha-Gal4 | F | 21051.77 | 0.59 |
| Cha-F-700181 | 4450 | ChaMARCM-F001492_seg001 | Cha-Gal4 | F | 18649.85 | 0.65 |
| Cha-F-000171 | 4946 | ChaMARCM-F001049_seg001 | Cha-Gal4 | F | 12431.39 | 0.56 |
| Cha-F-000175 | 4951 | ChaMARCM-F001051_seg004 | Cha-Gal4 | F | 24197.44 | 0.60 |
| Cha-F-800028 | 5378 | ChaMARCM-F000743_seg001 | Cha-Gal4 | F | 23910.51 | 0.69 |
| Cha-F-500127 | 7098 | ChaMARCM-F000574_seg002 | Cha-Gal4 | F | 20062.99 | 0.66 |
| Cha-F-400125 | 7119 | ChaMARCM-F000591_seg002 | Cha-Gal4 | F | 18515.76 | 0.63 |
| Cha-F-400092 | 7250 | ChaMARCM-F000422_seg001 | Cha-Gal4 | F | 15140.97 | 0.49 |
| Cha-F-800014 | 7531 | ChaMARCM-F000379_seg001 | Cha-Gal4 | F | 24648.36 | 0.74 |
| Cha-F-800024 | 7657 | ChaMARCM-F000488_seg001 | Cha-Gal4 | F | 24594.31 | 0.69 |
| Cha-F-500114 | 7735 | ChaMARCM-F000543_seg001 | Cha-Gal4 | F | 18298.73 | 0.62 |
| VGlut-F-500728 | 7959 | DvGlutMARCM-F003789_seg001 | VGlut-Gal4 | F | 26078.29 | 0.72 |
| Cha-F-400038 | 8195 | ChaMARCM-F000206_seg001 | Cha-Gal4 | F | 17017.48 | 0.58 |
| VGlut-F-600528 | 9335 | DvGlutMARCM-F003170_seg002 | VGlut-Gal4 | F | 19647.73 | 0.67 |
| npf-F-500001 | 9433 | npfMARCM-F000022_seg001 | npf-GAL4 | F | 21380.77 | 0.66 |
| npf-F-500007 | 9442 | npfMARCM-F000032_seg001 | npf-GAL4 | F | 15861.25 | 0.57 |
| npf-F-400003 | 9445 | npfMARCM-F000035_seg001 | npf-GAL4 | F | 20956.61 | 0.69 |
| npf-F-300015 | 9453 | npfMARCM-F000041_seg002 | npf-GAL4 | F | 20997.21 | 0.67 |
| npf-F-300016 | 9454 | npfMARCM-F000041_seg003 | npf-GAL4 | F | 19961.87 | 0.66 |
| Gad1-F-700013 | 10636 | GadMARCM-F000135_seg003 | Gad1-Gal4 | F | 11129.91 | 0.52 |
| Gad1-F-800019 | 11309 | GadMARCM-F000282_seg001 | Gad1-Gal4 | F | 16488.60 | 0.59 |
| Trh-F-000052 | 12594 | TPHMARCM-F001390_seg001 | Trh-Gal4 | F | 11234.62 | 0.50 |
| Trh-F-900023 | 13299 | TPHMARCM-781F_seg2 | Trh-Gal4 | F | 9581.18 | 0.46 |
| VGlut-F-100085 | 14328 | DvGlutMARCM-F1011_seg1 | VGlut-Gal4 | F | 15576.33 | 0.57 |
| VGlut-F-000250 | 14738 | DvGlutMARCM-F996_seg1 | VGlut-Gal4 | F | 26014.95 | 0.70 |
| VGlut-F-200213 | 14816 | DvGlutMARCM-F1161_seg1 | VGlut-Gal4 | F | 26080.11 | 0.70 |
| VGlut-F-100152 | 15076 | DvGlutMARCM-F1414_seg1 | VGlut-Gal4 | F | 24475.67 | 0.71 |
| VGlut-F-200279 | 15119 | DvGlutMARCM-F1452_seg1 | VGlut-Gal4 | F | 24092.42 | 0.73 |
| VGlut-F-100001 | 15635 | DvGlutMARCM-F048_seg1 | VGlut-Gal4 | F | 23364.94 | 0.72 |
| VGlut-F-000148 | 16168 | DvGlutMARCM-F549_seg1 | VGlut-Gal4 | F | 19835.33 | 0.64 |