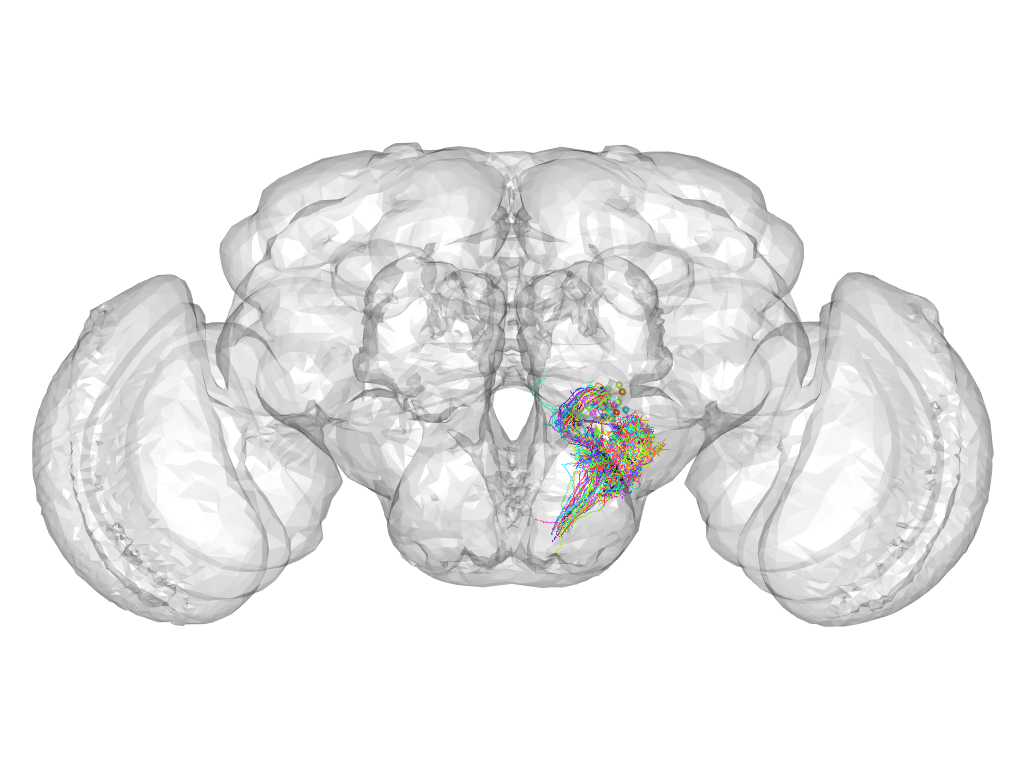
Cluster 432
Part of supercluster VII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-900133), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 528 | Cha-F-700078 | VII | 0.77 |
| 133 | fru-M-700043 | I | 0.64 |
| 860 | TH-F-300072 | VII | 0.62 |
| 991 | VGlut-F-400001 | VII | 0.58 |
| 660 | Tdc2-F-200049 | I | 0.56 |
| 397 | VGlut-F-200384 | VII | 0.56 |
| 776 | VGlut-F-500423 | IV | 0.52 |
| 916 | VGlut-F-300167 | VII | 0.52 |
Cluster composition
This cluster has 44 neurons. The exemplar of this cluster (VGlut-F-900133) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-900133 | 6668 | DvGlutMARCM-F004406_seg003 | VGlut-Gal4 | F | 9031.66 | 1.00 |
| Cha-F-300273 | 5038 | ChaMARCM-F001121_seg003 | Cha-Gal4 | F | 6650.04 | 0.73 |
| Cha-F-400235 | 5096 | ChaMARCM-F001157_seg001 | Cha-Gal4 | F | 6833.56 | 0.73 |
| Cha-F-200158 | 5403 | ChaMARCM-F000757_seg003 | Cha-Gal4 | F | 6656.56 | 0.71 |
| Cha-F-600116 | 5437 | ChaMARCM-F000780_seg003 | Cha-Gal4 | F | 6590.64 | 0.63 |
| Cha-F-800033 | 5463 | ChaMARCM-F000798_seg002 | Cha-Gal4 | F | 6671.26 | 0.71 |
| Cha-F-700156 | 5525 | ChaMARCM-F000834_seg001 | Cha-Gal4 | F | 6569.19 | 0.72 |
| Cha-F-700157 | 5526 | ChaMARCM-F000834_seg002 | Cha-Gal4 | F | 6451.21 | 0.74 |
| Cha-F-000150 | 5543 | ChaMARCM-F000845_seg003 | Cha-Gal4 | F | 6851.65 | 0.72 |
| Cha-F-500156 | 5571 | ChaMARCM-F000869_seg002 | Cha-Gal4 | F | 6573.08 | 0.70 |
| Cha-F-300245 | 5578 | ChaMARCM-F000873_seg002 | Cha-Gal4 | F | 6909.42 | 0.74 |
| Cha-F-600144 | 5628 | ChaMARCM-F000908_seg003 | Cha-Gal4 | F | 6511.57 | 0.68 |
| Cha-F-100118 | 5658 | ChaMARCM-F000928_seg002 | Cha-Gal4 | F | 6587.45 | 0.71 |
| Cha-F-800052 | 5679 | ChaMARCM-F000940_seg002 | Cha-Gal4 | F | 6909.17 | 0.75 |
| VGlut-F-700554 | 5711 | DvGlutMARCM-F004420_seg001 | VGlut-Gal4 | F | 6612.32 | 0.72 |
| VGlut-F-100389 | 5714 | DvGlutMARCM-F004422_seg002 | VGlut-Gal4 | F | 6992.53 | 0.77 |
| VGlut-F-200589 | 5716 | DvGlutMARCM-F004424_seg001 | VGlut-Gal4 | F | 6551.88 | 0.71 |
| VGlut-F-100394 | 5779 | DvGlutMARCM-F004483_seg001 | VGlut-Gal4 | F | 6891.96 | 0.74 |
| VGlut-F-800293 | 5792 | DvGlutMARCM-F004492_seg001 | VGlut-Gal4 | F | 6797.37 | 0.75 |
| VGlut-F-800294 | 5793 | DvGlutMARCM-F004492_seg002 | VGlut-Gal4 | F | 6913.80 | 0.76 |
| VGlut-F-500866 | 5796 | DvGlutMARCM-F004495_seg001 | VGlut-Gal4 | F | 6610.20 | 0.70 |
| VGlut-F-400904 | 5799 | DvGlutMARCM-F004497_seg002 | VGlut-Gal4 | F | 6491.58 | 0.66 |
| VGlut-F-000661 | 5841 | DvGlutMARCM-F004527_seg004 | VGlut-Gal4 | F | 6600.68 | 0.72 |
| VGlut-F-700583 | 5846 | DvGlutMARCM-F004532_seg001 | VGlut-Gal4 | F | 6791.79 | 0.75 |
| VGlut-F-000675 | 5870 | DvGlutMARCM-F004553_seg002 | VGlut-Gal4 | F | 6614.71 | 0.71 |
| VGlut-F-000677 | 5872 | DvGlutMARCM-F004554_seg002 | VGlut-Gal4 | F | 6511.24 | 0.72 |
| VGlut-F-300611 | 6439 | DvGlutMARCM-F004227_seg002 | VGlut-Gal4 | F | 6707.44 | 0.73 |
| VGlut-F-100372 | 6575 | DvGlutMARCM-F004337_seg001 | VGlut-Gal4 | F | 6971.68 | 0.75 |
| VGlut-F-100373 | 6576 | DvGlutMARCM-F004338_seg001 | VGlut-Gal4 | F | 6656.88 | 0.68 |
| VGlut-F-500821 | 6609 | DvGlutMARCM-F004365_seg001 | VGlut-Gal4 | F | 6892.80 | 0.75 |
| VGlut-F-700544 | 6627 | DvGlutMARCM-F004378_seg002 | VGlut-Gal4 | F | 6710.64 | 0.71 |
| VGlut-F-200578 | 6647 | DvGlutMARCM-F004392_seg003 | VGlut-Gal4 | F | 6535.67 | 0.71 |
| VGlut-F-100383 | 6672 | DvGlutMARCM-F004410_seg002 | VGlut-Gal4 | F | 6774.56 | 0.74 |
| VGlut-F-100384 | 6673 | DvGlutMARCM-F004410_seg003 | VGlut-Gal4 | F | 6617.66 | 0.71 |
| Cha-F-300163 | 7142 | ChaMARCM-F000607_seg003 | Cha-Gal4 | F | 6555.74 | 0.71 |
| Cha-F-600045 | 7219 | ChaMARCM-F000388_seg001 | Cha-Gal4 | F | 6418.66 | 0.68 |
| Cha-F-300118 | 7496 | ChaMARCM-F000360_seg002 | Cha-Gal4 | F | 6651.72 | 0.73 |
| Cha-F-600052 | 7540 | ChaMARCM-F000393_seg004 | Cha-Gal4 | F | 6671.37 | 0.71 |
| Cha-F-700100 | 7558 | ChaMARCM-F000407_seg003 | Cha-Gal4 | F | 6600.34 | 0.68 |
| Cha-F-200006 | 10073 | ChaMARCM-F000034_seg002 | Cha-Gal4 | F | 6388.28 | 0.71 |
| Cha-F-400028 | 10130 | ChaMARCM-F000076_seg003 | Cha-Gal4 | F | 6596.26 | 0.74 |
| VGlut-F-100027 | 15927 | DvGlutMARCM-F322_seg1 | VGlut-Gal4 | F | 6769.17 | 0.73 |
| VGlut-F-000175 | 16206 | DvGlutMARCM-F586_seg1 | VGlut-Gal4 | F | 6327.94 | 0.68 |
| VGlut-F-000178 | 16215 | DvGlutMARCM-F593_seg1 | VGlut-Gal4 | F | 6634.00 | 0.75 |