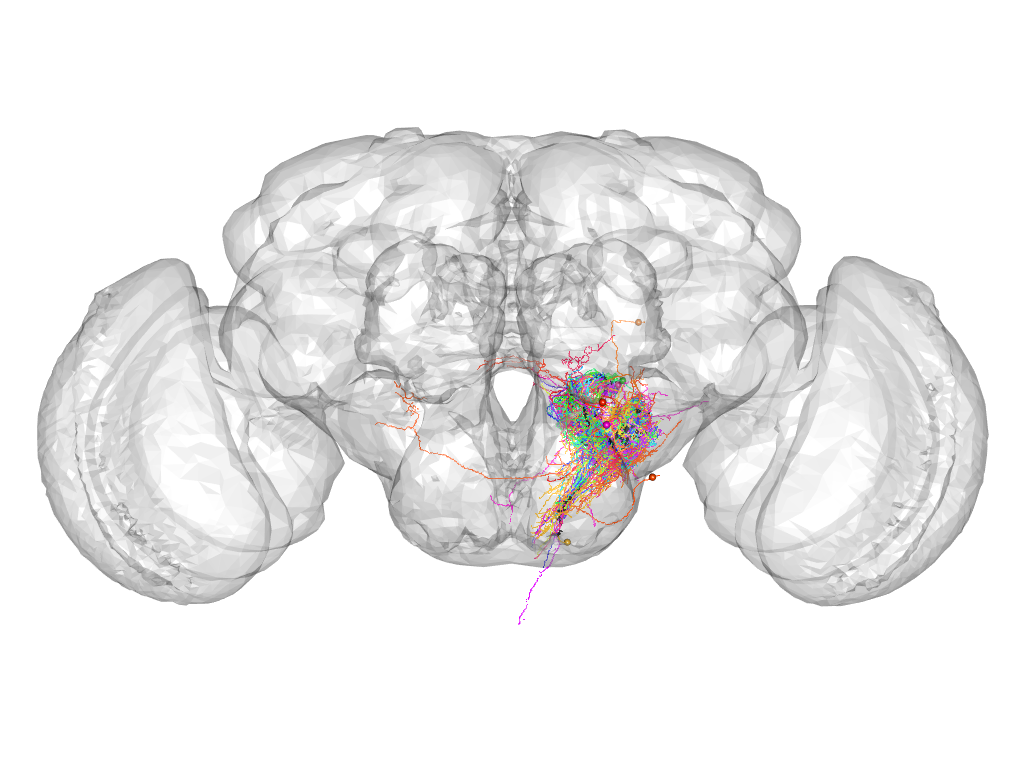
Cluster 528
Part of supercluster VII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Cha-F-700078), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 432 | VGlut-F-900133 | VII | 0.70 |
| 133 | fru-M-700043 | I | 0.64 |
| 860 | TH-F-300072 | VII | 0.58 |
| 916 | VGlut-F-300167 | VII | 0.58 |
| 397 | VGlut-F-200384 | VII | 0.56 |
| 991 | VGlut-F-400001 | VII | 0.55 |
| 660 | Tdc2-F-200049 | I | 0.54 |
| 776 | VGlut-F-500423 | IV | 0.52 |
Cluster composition
This cluster has 44 neurons. The exemplar of this cluster (Cha-F-700078) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Cha-F-700078 | 8250 | ChaMARCM-F000239_seg003 | Cha-Gal4 | F | 26479.96 | 1.00 |
| Cha-F-300336 | 3900 | ChaMARCM-F001562_seg001 | Cha-Gal4 | F | 16921.91 | 0.63 |
| Gad1-F-100070 | 3984 | GadMARCM-F000605_seg002 | Gad1-Gal4 | F | 17094.83 | 0.52 |
| Gad1-F-000060 | 3988 | GadMARCM-F000608_seg002 | Gad1-Gal4 | F | 14311.45 | 0.35 |
| Gad1-F-800072 | 4012 | GadMARCM-F000625_seg002 | Gad1-Gal4 | F | 16534.81 | 0.55 |
| Cha-F-600167 | 4200 | ChaMARCM-F001296_seg002 | Cha-Gal4 | F | 18154.93 | 0.72 |
| Cha-F-100161 | 4906 | ChaMARCM-F001019_seg001 | Cha-Gal4 | F | 14173.14 | 0.44 |
| Cha-F-300274 | 5039 | ChaMARCM-F001121_seg004 | Cha-Gal4 | F | 18916.11 | 0.71 |
| Cha-F-000121 | 5342 | ChaMARCM-F000714_seg002 | Cha-Gal4 | F | 18698.00 | 0.70 |
| Cha-F-800029 | 5405 | ChaMARCM-F000759_seg001 | Cha-Gal4 | F | 19694.41 | 0.72 |
| Cha-F-300220 | 5471 | ChaMARCM-F000801_seg001 | Cha-Gal4 | F | 19114.58 | 0.70 |
| Cha-F-700151 | 5520 | ChaMARCM-F000832_seg002 | Cha-Gal4 | F | 18942.90 | 0.74 |
| Cha-F-700155 | 5524 | ChaMARCM-F000833_seg004 | Cha-Gal4 | F | 19038.50 | 0.71 |
| Cha-F-200164 | 5532 | ChaMARCM-F000840_seg001 | Cha-Gal4 | F | 18231.62 | 0.72 |
| Cha-F-000149 | 5542 | ChaMARCM-F000845_seg002 | Cha-Gal4 | F | 18047.49 | 0.68 |
| Cha-F-200173 | 5557 | ChaMARCM-F000857_seg001 | Cha-Gal4 | F | 18355.84 | 0.72 |
| Cha-F-000156 | 5584 | ChaMARCM-F000877_seg002 | Cha-Gal4 | F | 18804.77 | 0.71 |
| Cha-F-100113 | 5651 | ChaMARCM-F000924_seg004 | Cha-Gal4 | F | 19262.93 | 0.70 |
| Cha-F-100114 | 5652 | ChaMARCM-F000924_seg005 | Cha-Gal4 | F | 19095.11 | 0.73 |
| Cha-F-300255 | 5669 | ChaMARCM-F000936_seg001 | Cha-Gal4 | F | 19598.23 | 0.74 |
| Cha-F-300256 | 5670 | ChaMARCM-F000936_seg002 | Cha-Gal4 | F | 18946.60 | 0.74 |
| Cha-F-300263 | 5681 | ChaMARCM-F000941_seg002 | Cha-Gal4 | F | 19586.67 | 0.74 |
| Cha-F-500169 | 5683 | ChaMARCM-F000942_seg002 | Cha-Gal4 | F | 18782.24 | 0.70 |
| Cha-F-200002 | 6136 | ChaMARCM-F000004_seg005 | Cha-Gal4 | F | 18754.89 | 0.71 |
| Cha-F-500126 | 7097 | ChaMARCM-F000574_seg001 | Cha-Gal4 | F | 18546.12 | 0.68 |
| Cha-F-300155 | 7130 | ChaMARCM-F000598_seg002 | Cha-Gal4 | F | 18520.82 | 0.71 |
| Cha-F-700134 | 7261 | ChaMARCM-F000554_seg001 | Cha-Gal4 | F | 18135.93 | 0.69 |
| Cha-F-700118 | 7602 | ChaMARCM-F000450_seg003 | Cha-Gal4 | F | 18917.65 | 0.72 |
| Cha-F-400108 | 7610 | ChaMARCM-F000455_seg001 | Cha-Gal4 | F | 18465.06 | 0.69 |
| Cha-F-200086 | 7638 | ChaMARCM-F000473_seg002 | Cha-Gal4 | F | 19514.77 | 0.72 |
| Cha-F-800021 | 7650 | ChaMARCM-F000482_seg001 | Cha-Gal4 | F | 18364.64 | 0.68 |
| Cha-F-400113 | 7662 | ChaMARCM-F000489_seg004 | Cha-Gal4 | F | 17434.59 | 0.67 |
| Cha-F-000081 | 7689 | ChaMARCM-F000511_seg001 | Cha-Gal4 | F | 18958.56 | 0.73 |
| Cha-F-000083 | 7691 | ChaMARCM-F000512_seg001 | Cha-Gal4 | F | 19024.11 | 0.72 |
| Cha-F-600082 | 7722 | ChaMARCM-F000534_seg001 | Cha-Gal4 | F | 17895.09 | 0.71 |
| Cha-F-700131 | 7744 | ChaMARCM-F000548_seg002 | Cha-Gal4 | F | 18355.17 | 0.61 |
| Cha-F-700077 | 8249 | ChaMARCM-F000239_seg002 | Cha-Gal4 | F | 20646.33 | 0.75 |
| Cha-F-600024 | 8341 | ChaMARCM-F000292_seg002 | Cha-Gal4 | F | 17251.66 | 0.65 |
| Cha-F-000059 | 8350 | ChaMARCM-F000300_seg002 | Cha-Gal4 | F | 16384.59 | 0.48 |
| Cha-F-100011 | 10090 | ChaMARCM-F000050_seg003 | Cha-Gal4 | F | 18759.91 | 0.73 |
| TH-F-300011 | 13388 | THMARCM-156F_seg1 | TH-Gal4 | F | 19203.82 | 0.71 |
| TH-F-300040 | 13469 | THMARCM-304F_seg1 | TH-Gal4 | F | 19481.96 | 0.72 |
| VGlut-F-000320 | 14848 | DvGlutMARCM-F1195_seg1 | VGlut-Gal4 | F | 18095.54 | 0.56 |
| VGlut-F-500081 | 16051 | DvGlutMARCM-F440_seg2 | VGlut-Gal4 | F | 17124.61 | 0.69 |