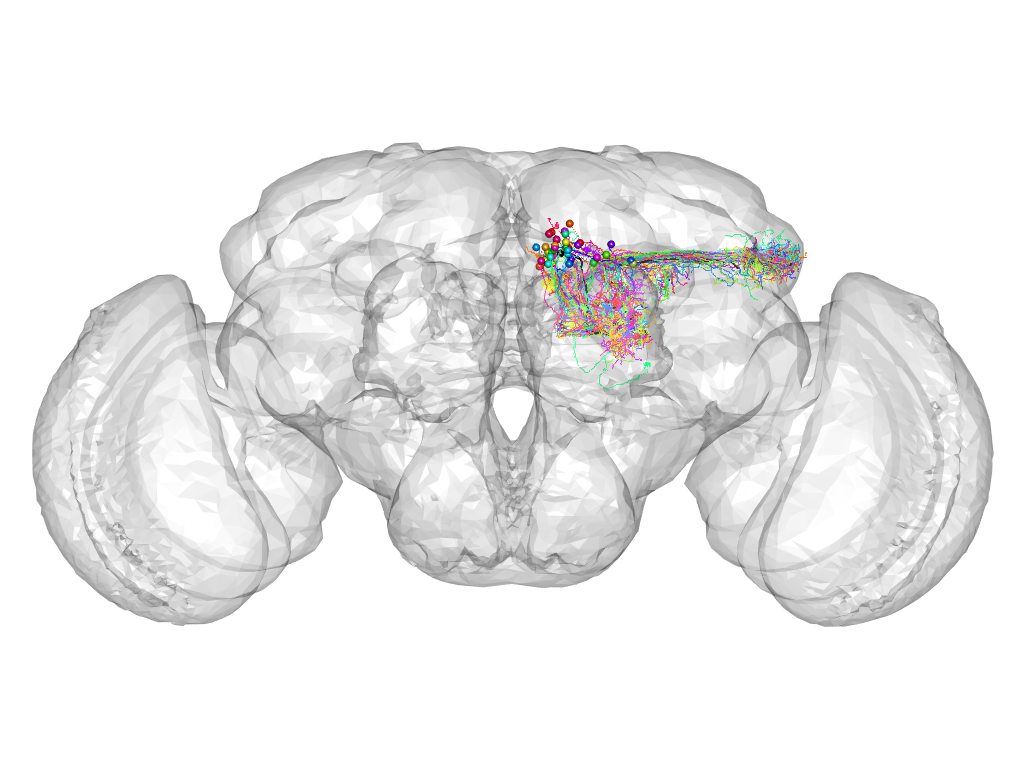
Cluster 384
Part of supercluster XI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-600797), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 662 | Tdc2-F-400026 | XIII | 0.70 |
| 549 | VGlut-F-500582 | XI | 0.59 |
| 815 | VGlut-F-400462 | XI | 0.52 |
| 54 | fru-M-500169 | XI | 0.48 |
| 458 | VGlut-F-700500 | XI | 0.45 |
Cluster composition
This cluster has 40 neurons. The exemplar of this cluster (VGlut-F-600797) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-600797 | 5962 | DvGlutMARCM-F004613_seg001 | VGlut-Gal4 | F | 11161.45 | 1.00 |
| VGlut-F-300646 | 5735 | DvGlutMARCM-F004441_seg001 | VGlut-Gal4 | F | 7258.27 | 0.63 |
| Trh-F-000062 | 6403 | TPHMARCM-F001824_seg001 | Trh-Gal4 | F | 7952.07 | 0.61 |
| VGlut-F-200560 | 6530 | DvGlutMARCM-F004300_seg001 | VGlut-Gal4 | F | 7369.97 | 0.64 |
| VGlut-F-300641 | 6589 | DvGlutMARCM-F004349_seg001 | VGlut-Gal4 | F | 7778.89 | 0.66 |
| VGlut-F-600770 | 7040 | DvGlutMARCM-F004173_seg002 | VGlut-Gal4 | F | 7979.42 | 0.63 |
| VGlut-F-700504 | 7045 | DvGlutMARCM-F004176_seg001 | VGlut-Gal4 | F | 8026.85 | 0.69 |
| Cha-F-100073 | 7156 | ChaMARCM-F000615_seg001 | Cha-Gal4 | F | 7226.62 | 0.64 |
| VGlut-F-500407 | 8420 | DvGlutMARCM-F002507_seg002 | VGlut-Gal4 | F | 7765.14 | 0.58 |
| VGlut-F-500497 | 8422 | DvGlutMARCM-F002923_seg001 | VGlut-Gal4 | F | 7146.87 | 0.60 |
| VGlut-F-400662 | 8452 | DvGlutMARCM-F003051_seg001 | VGlut-Gal4 | F | 5192.57 | 0.45 |
| VGlut-F-700287 | 8524 | DvGlutMARCM-F003103_seg001 | VGlut-Gal4 | F | 7799.40 | 0.69 |
| VGlut-F-400719 | 8966 | DvGlutMARCM-F003524_seg001 | VGlut-Gal4 | F | 7294.50 | 0.66 |
| VGlut-F-700323 | 9269 | DvGlutMARCM-F003327_seg001 | VGlut-Gal4 | F | 7364.27 | 0.66 |
| VGlut-F-400668 | 9360 | DvGlutMARCM-F003194_seg001 | VGlut-Gal4 | F | 7183.38 | 0.62 |
| VGlut-F-500594 | 9375 | DvGlutMARCM-F003206_seg001 | VGlut-Gal4 | F | 7957.11 | 0.70 |
| Gad1-F-400075 | 9521 | GadMARCM-F000406_seg002 | Gad1-Gal4 | F | 8071.72 | 0.73 |
| fru-F-000025 | 9986 | FruMARCM-F000575_seg001 | fru-Gal4 | F | 5622.45 | 0.44 |
| VGlut-F-700124 | 10850 | DvGlutMARCM-F002776_seg001 | VGlut-Gal4 | F | 7171.55 | 0.63 |
| VGlut-F-700147 | 10872 | DvGlutMARCM-F002784_seg001 | VGlut-Gal4 | F | 7836.91 | 0.67 |
| VGlut-F-700185 | 10909 | DvGlutMARCM-F002810_seg001 | VGlut-Gal4 | F | 7396.33 | 0.67 |
| VGlut-F-800089 | 10977 | DvGlutMARCM-F002867_seg001 | VGlut-Gal4 | F | 7768.58 | 0.70 |
| VGlut-F-600471 | 11115 | DvGlutMARCM-F002981_seg001 | VGlut-Gal4 | F | 7924.38 | 0.68 |
| VGlut-F-600482 | 11164 | DvGlutMARCM-F003013_seg001 | VGlut-Gal4 | F | 7569.20 | 0.68 |
| VGlut-F-800047 | 12076 | DvGlutMARCM-F002585_seg001 | VGlut-Gal4 | F | 7446.95 | 0.59 |
| VGlut-F-600327 | 12156 | DvGlutMARCM-F002646_seg001 | VGlut-Gal4 | F | 7388.68 | 0.67 |
| VGlut-F-600353 | 12183 | DvGlutMARCM-F002660_seg001 | VGlut-Gal4 | F | 7928.99 | 0.70 |
| VGlut-F-700107 | 12247 | DvGlutMARCM-F002710_seg001 | VGlut-Gal4 | F | 7652.58 | 0.65 |
| VGlut-F-400432 | 12772 | DvGlutMARCM-F1827_seg1 | VGlut-Gal4 | F | 7340.27 | 0.66 |
| VGlut-F-500303 | 12877 | DvGlutMARCM-F1918_seg1 | VGlut-Gal4 | F | 7690.41 | 0.70 |
| VGlut-F-500333 | 13045 | DvGlutMARCM-F2068_seg1 | VGlut-Gal4 | F | 7419.29 | 0.66 |
| VGlut-F-400218 | 14517 | DvGlutMARCM-F788-x2_seg2 | VGlut-Gal4 | F | 7449.93 | 0.66 |
| VGlut-F-500118 | 14603 | DvGlutMARCM-F865_seg1 | VGlut-Gal4 | F | 7891.13 | 0.65 |
| VGlut-F-400376 | 15085 | DvGlutMARCM-F1421_seg1 | VGlut-Gal4 | F | 8093.39 | 0.61 |
| VGlut-F-500189 | 15224 | DvGlutMARCM-F1548_seg2 | VGlut-Gal4 | F | 7989.29 | 0.71 |
| VGlut-F-000391 | 15289 | DvGlutMARCM-F1620_seg1 | VGlut-Gal4 | F | 7066.72 | 0.63 |
| VGlut-F-400402 | 15318 | DvGlutMARCM-F1658_seg1 | VGlut-Gal4 | F | 7154.97 | 0.62 |
| VGlut-F-400407 | 15331 | DvGlutMARCM-F1675_seg1 | VGlut-Gal4 | F | 7585.37 | 0.63 |
| VGlut-F-500237 | 15361 | DvGlutMARCM-F1714_seg1 | VGlut-Gal4 | F | 7268.97 | 0.65 |
| VGlut-F-500076 | 16039 | DvGlutMARCM-F429_seg1 | VGlut-Gal4 | F | 7440.22 | 0.65 |