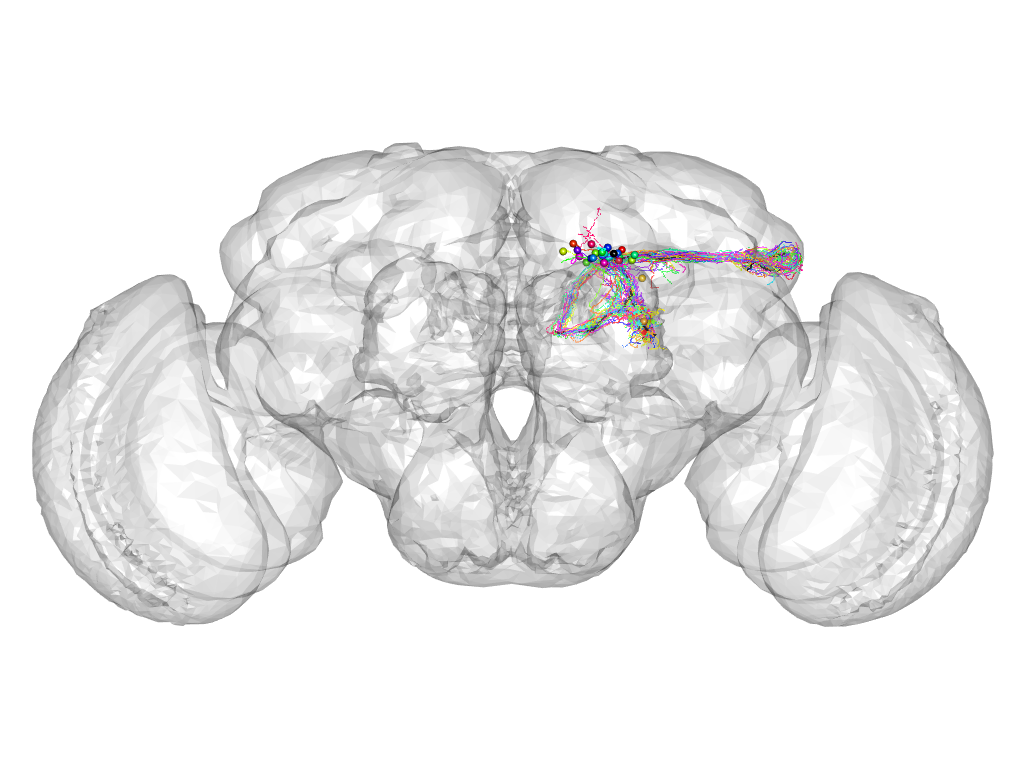
Cluster 54
Part of supercluster XI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-500169), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 815 | VGlut-F-400462 | XI | 0.73 |
| 549 | VGlut-F-500582 | XI | 0.72 |
| 384 | VGlut-F-600797 | XI | 0.59 |
| 662 | Tdc2-F-400026 | XIII | 0.57 |
| 413 | fru-F-400149 | XII | 0.43 |
Cluster composition
This cluster has 32 neurons. The exemplar of this cluster (fru-M-500169) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-500169 | 593 | FruMARCM-M001786_seg002 | fru-Gal4 | M | 7608.01 | 1.00 |
| fru-F-000079 | 4524 | FruMARCM-F001637_seg001 | fru-Gal4 | F | 5675.74 | 0.73 |
| VGlut-F-600798 | 5963 | DvGlutMARCM-F004614_seg001 | VGlut-Gal4 | F | 5732.96 | 0.73 |
| VGlut-F-600390 | 6037 | DvGlutMARCM-F002707_seg001 | VGlut-Gal4 | F | 5791.72 | 0.75 |
| VGlut-F-900127 | 6506 | DvGlutMARCM-F004285_seg001 | VGlut-Gal4 | F | 5444.07 | 0.73 |
| VGlut-F-700539 | 6577 | DvGlutMARCM-F004339_seg001 | VGlut-Gal4 | F | 5478.30 | 0.72 |
| VGlut-F-500826 | 6614 | DvGlutMARCM-F004367_seg001 | VGlut-Gal4 | F | 5748.98 | 0.73 |
| VGlut-F-600751 | 6929 | DvGlutMARCM-F004084_seg001 | VGlut-Gal4 | F | 5705.52 | 0.72 |
| VGlut-F-500772 | 6988 | DvGlutMARCM-F004134_seg001 | VGlut-Gal4 | F | 5767.76 | 0.76 |
| VGlut-F-500548 | 8489 | DvGlutMARCM-F003077_seg001 | VGlut-Gal4 | F | 5714.22 | 0.74 |
| VGlut-F-500638 | 9281 | DvGlutMARCM-F003333_seg001 | VGlut-Gal4 | F | 5730.46 | 0.75 |
| VGlut-F-300458 | 9313 | DvGlutMARCM-F003154_seg001 | VGlut-Gal4 | F | 5757.78 | 0.73 |
| VGlut-F-700123 | 10849 | DvGlutMARCM-F002775_seg002 | VGlut-Gal4 | F | 5469.00 | 0.70 |
| VGlut-F-500502 | 11143 | DvGlutMARCM-F003000_seg001 | VGlut-Gal4 | F | 5667.77 | 0.70 |
| VGlut-F-600494 | 11175 | DvGlutMARCM-F003022_seg001 | VGlut-Gal4 | F | 5451.52 | 0.72 |
| VGlut-F-800121 | 11561 | DvGlutMARCM-F003036_seg001 | VGlut-Gal4 | F | 5712.85 | 0.76 |
| VGlut-F-800123 | 11563 | DvGlutMARCM-F003037_seg001 | VGlut-Gal4 | F | 5788.54 | 0.73 |
| VGlut-F-700245 | 11801 | DvGlutMARCM-F002366_seg001 | VGlut-Gal4 | F | 5549.50 | 0.73 |
| VGlut-F-400531 | 11883 | DvGlutMARCM-F002424_seg002 | VGlut-Gal4 | F | 5430.20 | 0.73 |
| VGlut-F-600314 | 12056 | DvGlutMARCM-F002570_seg001 | VGlut-Gal4 | F | 5604.06 | 0.74 |
| VGlut-F-600371 | 12208 | DvGlutMARCM-F002676_seg001 | VGlut-Gal4 | F | 5755.80 | 0.76 |
| VGlut-F-600375 | 12221 | DvGlutMARCM-F002688_seg001 | VGlut-Gal4 | F | 5719.58 | 0.77 |
| VGlut-F-100221 | 12331 | DvGlutMARCM-F2112_seg1 | VGlut-Gal4 | F | 5678.17 | 0.73 |
| VGlut-F-400444 | 12828 | DvGlutMARCM-F1876_seg1 | VGlut-Gal4 | F | 5641.09 | 0.71 |
| VGlut-F-400280 | 14677 | DvGlutMARCM-F938_seg1 | VGlut-Gal4 | F | 5689.57 | 0.73 |
| VGlut-F-500169 | 15168 | DvGlutMARCM-F1500_seg1 | VGlut-Gal4 | F | 5274.03 | 0.70 |
| VGlut-F-500211 | 15292 | DvGlutMARCM-F1624_seg1 | VGlut-Gal4 | F | 5890.55 | 0.72 |
| VGlut-F-500259 | 15426 | DvGlutMARCM-F1782_seg1 | VGlut-Gal4 | F | 5776.71 | 0.74 |
| VGlut-F-400023 | 15713 | DvGlutMARCM-F126_seg1 | VGlut-Gal4 | F | 5424.47 | 0.68 |
| VGlut-F-700008 | 15736 | DvGlutMARCM-F147_seg1 | VGlut-Gal4 | F | 5932.42 | 0.77 |
| VGlut-F-600006 | 15818 | DvGlutMARCM-F224-R_seg2 | VGlut-Gal4 | F | 5331.34 | 0.63 |
| VGlut-F-600016 | 15881 | DvGlutMARCM-F281-X2_seg2 | VGlut-Gal4 | F | 5675.82 | 0.75 |