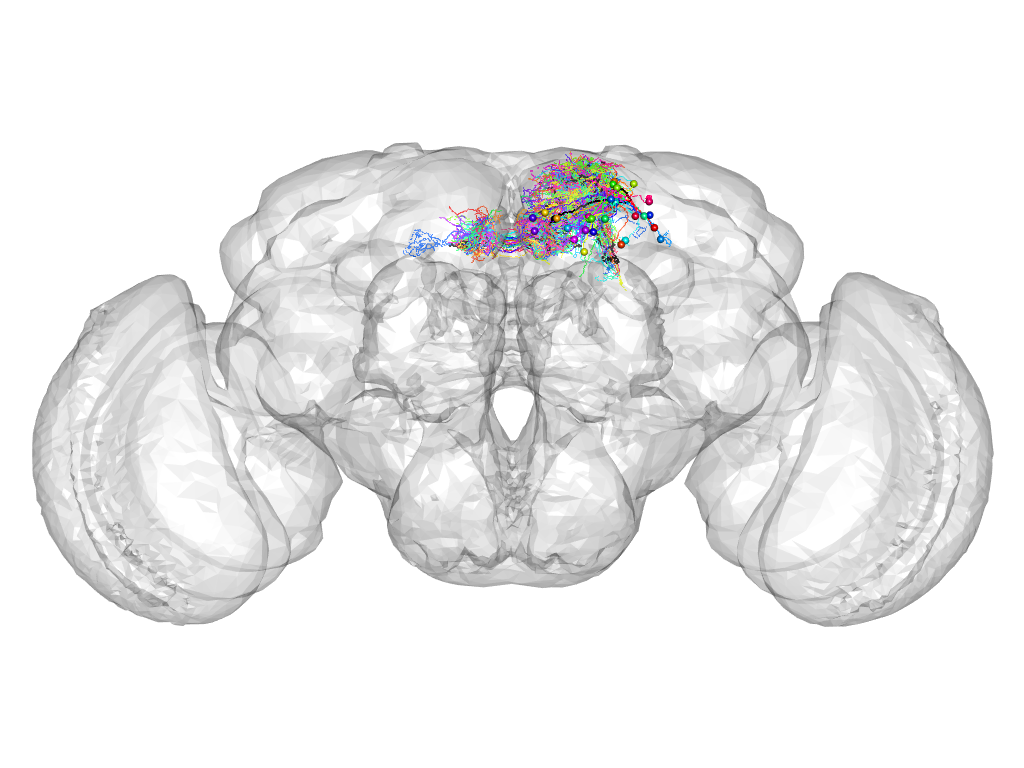
Cluster 186
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-M-600001), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 867 | TH-F-100077 | XIX | 0.65 |
| 638 | fru-F-400079 | XIX | 0.52 |
| 197 | TH-M-200095 | XIX | 0.49 |
| 204 | Trh-M-100025 | XIV | 0.49 |
| 941 | VGlut-F-200251 | XIX | 0.48 |
Cluster composition
This cluster has 29 neurons. The exemplar of this cluster (TH-M-600001) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-M-600001 | 3125 | THMARCM-163M_seg1 | TH-Gal4 | M | 24874.08 | 1.00 |
| fru-M-400209 | 128 | FruMARCM-M002324_seg001 | fru-Gal4 | M | 12465.81 | 0.55 |
| fru-M-500174 | 650 | FruMARCM-M001861_seg001 | fru-Gal4 | M | 14257.10 | 0.60 |
| fru-M-400114 | 976 | FruMARCM-M001150_seg001 | fru-Gal4 | M | 12610.69 | 0.59 |
| fru-M-500026 | 1946 | FruMARCM-M000540_seg003 | fru-Gal4 | M | 12455.14 | 0.59 |
| Trh-M-400088 | 2770 | TPHMARCM-1116M_seg1 | Trh-Gal4 | M | 13173.88 | 0.59 |
| TH-M-700018 | 3049 | THMARCM-148M_seg1 | TH-Gal4 | M | 15343.36 | 0.63 |
| TH-M-700005 | 3108 | THMARCM-121M_seg1 | TH-Gal4 | M | 13168.03 | 0.61 |
| TH-M-700009 | 3112 | THMARCM-126M_seg1 | TH-Gal4 | M | 13641.55 | 0.58 |
| TH-M-700013 | 3115 | THMARCM-129M_seg1 | TH-Gal4 | M | 15232.84 | 0.66 |
| TH-M-700014 | 3116 | THMARCM-133M_seg1 | TH-Gal4 | M | 13946.09 | 0.63 |
| TH-M-600002 | 3133 | THMARCM-173M_seg1 | TH-Gal4 | M | 15045.24 | 0.64 |
| TH-M-600003 | 3339 | THMARCM-625M_seg | TH-Gal4 | M | 12170.51 | 0.55 |
| fru-F-500263 | 3717 | FruMARCM-F002212_seg001 | fru-Gal4 | F | 14887.88 | 0.64 |
| fru-F-500235 | 4710 | FruMARCM-F002038_seg001 | fru-Gal4 | F | 15629.79 | 0.65 |
| fru-F-600114 | 4713 | FruMARCM-F002041_seg001 | fru-Gal4 | F | 14661.65 | 0.62 |
| fru-F-500240 | 4731 | FruMARCM-F002057_seg002 | fru-Gal4 | F | 14195.67 | 0.62 |
| fru-F-500254 | 4783 | FruMARCM-F002104_seg003 | fru-Gal4 | F | 15433.48 | 0.63 |
| VGlut-F-500070 | 6124 | DvGlutMARCM-F423-X3_seg1 | VGlut-Gal4 | F | 10137.35 | 0.45 |
| fru-F-700114 | 6177 | FruMARCM-F001111_seg001 | fru-Gal4 | F | 14687.59 | 0.64 |
| fru-F-700063 | 7270 | FruMARCM-F000908_seg001 | fru-Gal4 | F | 14447.71 | 0.64 |
| Cha-F-400112 | 7661 | ChaMARCM-F000489_seg003 | Cha-Gal4 | F | 11516.01 | 0.50 |
| fru-F-500143 | 7827 | FruMARCM-F000720_seg001 | fru-Gal4 | F | 12923.48 | 0.57 |
| fru-F-500011 | 9839 | FruMARCM-F000406_seg001 | fru-Gal4 | F | 15397.77 | 0.63 |
| fru-F-600021 | 9940 | FruMARCM-F000526_seg001 | fru-Gal4 | F | 13193.43 | 0.61 |
| TH-F-100008 | 13338 | THMARCM-071F_seg1 | TH-Gal4 | F | 14707.29 | 0.62 |
| TH-F-700016 | 13362 | THMARCM-106_seg1 | TH-Gal4 | F | 15616.30 | 0.67 |
| TH-F-700017 | 13363 | THMARCM-107_seg1 | TH-Gal4 | F | 15489.31 | 0.65 |
| Trh-F-300031 | 14003 | TPHMARCM-293F_seg1 | Trh-Gal4 | F | 13312.20 | 0.61 |