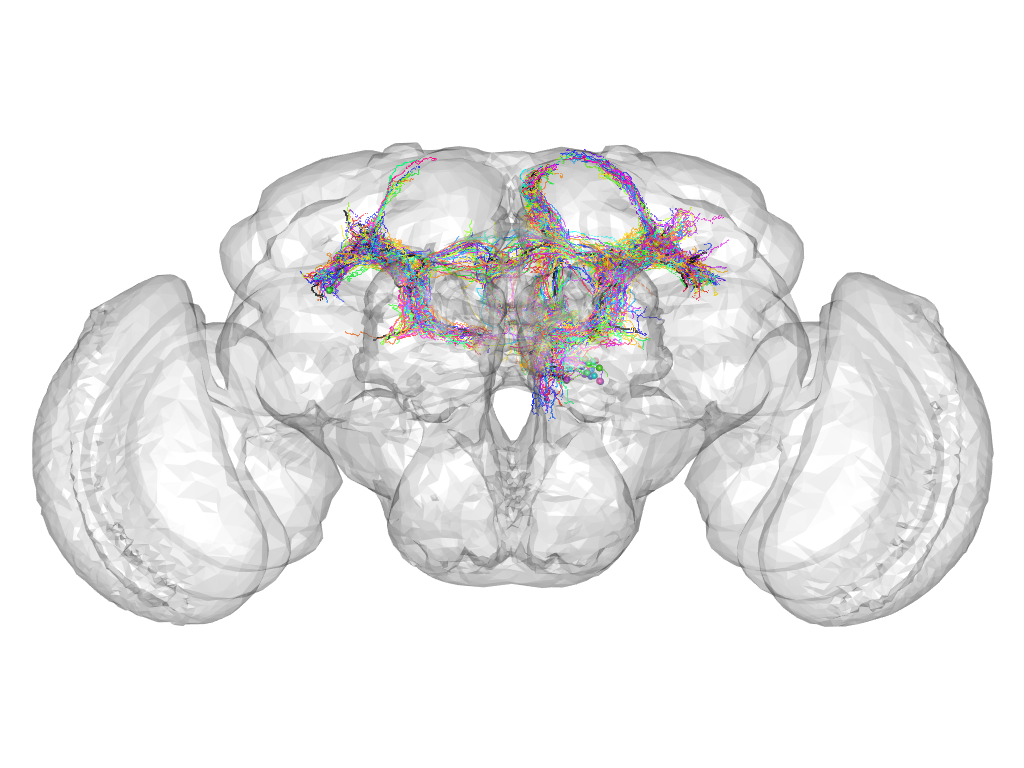
Cluster 885
Part of supercluster XVIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-200059), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 165 | Trh-M-500167 | XVIII | 0.57 |
| 411 | fru-F-700151 | X | 0.24 |
| 33 | fru-M-400224 | X | 0.24 |
| 306 | fru-F-600108 | X | 0.20 |
| 30 | fru-M-000203 | XVIII | 0.19 |
Cluster composition
This cluster has 35 neurons. The exemplar of this cluster (Trh-F-200059) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-200059 | 14022 | TPHMARCM-309F_seg1 | Trh-Gal4 | F | 29520.88 | 1.00 |
| Trh-M-000156 | 936 | TPHMARCM-M001842_seg001 | Trh-Gal4 | M | 20112.93 | 0.67 |
| Trh-M-000158 | 938 | TPHMARCM-M001843_seg001 | Trh-Gal4 | M | 19897.74 | 0.68 |
| Trh-M-000159 | 939 | TPHMARCM-M001844_seg001 | Trh-Gal4 | M | 19269.58 | 0.64 |
| fru-M-600047 | 1051 | FruMARCM-M001231_seg001 | fru-Gal4 | M | 16598.73 | 0.57 |
| Trh-M-000087 | 2259 | TPHMARCM-M001746_seg001 | Trh-Gal4 | M | 17907.88 | 0.60 |
| Trh-M-000109 | 2295 | TPHMARCM-M001777_seg001 | Trh-Gal4 | M | 16526.46 | 0.55 |
| fru-F-000124 | 3751 | FruMARCM-F002342_seg001 | fru-Gal4 | F | 21085.10 | 0.69 |
| fru-F-000142 | 3824 | FruMARCM-F002635_seg001 | fru-Gal4 | F | 20648.24 | 0.68 |
| fru-F-200077 | 6218 | FruMARCM-F001141_seg001 | fru-Gal4 | F | 19301.01 | 0.67 |
| fru-F-200081 | 6245 | FruMARCM-F001256_seg001 | fru-Gal4 | F | 20246.06 | 0.70 |
| fru-F-000047 | 6275 | FruMARCM-F001306_seg001 | fru-Gal4 | F | 19936.97 | 0.68 |
| fru-F-000052 | 6291 | FruMARCM-F001387_seg001 | fru-Gal4 | F | 19116.19 | 0.66 |
| fru-F-000066 | 6387 | FruMARCM-F001524_seg001 | fru-Gal4 | F | 16742.85 | 0.59 |
| fru-F-600053 | 7308 | FruMARCM-F000948_seg001 | fru-Gal4 | F | 18405.81 | 0.65 |
| fru-F-300049 | 7892 | FruMARCM-F000823_seg001 | fru-Gal4 | F | 19033.36 | 0.62 |
| Gad1-F-600038 | 11360 | GadMARCM-F000318_seg001 | Gad1-Gal4 | F | 18540.25 | 0.66 |
| Trh-F-100073 | 12493 | TPHMARCM-1279F_seg1 | Trh-Gal4 | F | 19457.86 | 0.68 |
| Trh-F-200132 | 12572 | TPHMARCM-F001371_seg001 | Trh-Gal4 | F | 18516.43 | 0.65 |
| Trh-F-000050 | 12592 | TPHMARCM-F001388_seg001 | Trh-Gal4 | F | 18274.82 | 0.65 |
| Trh-F-000054 | 12596 | TPHMARCM-F001392_seg001 | Trh-Gal4 | F | 20619.22 | 0.68 |
| Trh-F-100089 | 12603 | TPHMARCM-F001398_seg001 | Trh-Gal4 | F | 18557.75 | 0.67 |
| Trh-F-000058 | 12612 | TPHMARCM-F001435_seg001 | Trh-Gal4 | F | 21131.63 | 0.68 |
| VGlut-F-500314 | 12742 | DVGlutMARCM-F2003-montag_seg1 | VGlut-Gal4 | F | 20072.95 | 0.65 |
| Trh-F-300111 | 13303 | TPHMARCM-785F_seg1 | Trh-Gal4 | F | 19249.34 | 0.64 |
| Trh-F-100022 | 13815 | TPHMARCM-034F_seg1 | Trh-Gal4 | F | 21033.89 | 0.70 |
| Trh-F-100033 | 13911 | TPHMARCM-179F_seg1 | Trh-Gal4 | F | 20447.33 | 0.70 |
| Trh-F-200017 | 13938 | TPHMARCM-230F_seg1 | Trh-Gal4 | F | 19354.17 | 0.65 |
| Trh-F-000023 | 13946 | TPHMARCM-238F_seg1 | Trh-Gal4 | F | 19883.67 | 0.67 |
| Trh-F-200027 | 13955 | TPHMARCM-246F_seg1 | Trh-Gal4 | F | 19553.11 | 0.65 |
| Trh-F-300022 | 13976 | TPHMARCM-266F_seg1 | Trh-Gal4 | F | 19821.01 | 0.66 |
| Trh-F-300051 | 14078 | TPHMARCM-427F_seg1 | Trh-Gal4 | F | 19636.95 | 0.67 |
| Trh-F-300085 | 14173 | TPHMARCM-610F_seg2 | Trh-Gal4 | F | 20250.95 | 0.67 |
| Trh-F-400065 | 14206 | TPHMARCM-647F_seg1 | Trh-Gal4 | F | 19833.84 | 0.67 |
| Trh-F-200082 | 14211 | TPHMARCM-651F_seg1 | Trh-Gal4 | F | 19616.23 | 0.66 |