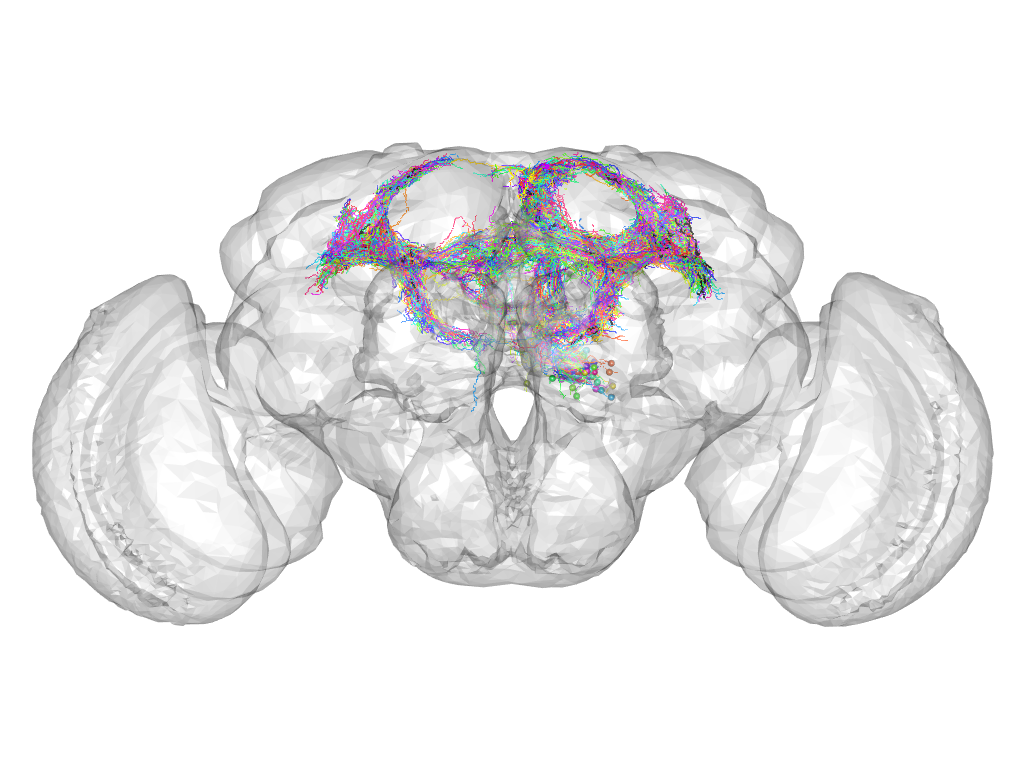
Cluster 165
Part of supercluster XVIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-500167), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 885 | Trh-F-200059 | XVIII | 0.57 |
| 30 | fru-M-000203 | XVIII | 0.50 |
| 203 | Trh-M-000010 | XVIII | 0.46 |
| 1025 | VGlut-F-000057 | XVIII | 0.35 |
| 22 | fru-M-300288 | XVIII | 0.33 |
Cluster composition
This cluster has 75 neurons. The exemplar of this cluster (Trh-M-500167) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-500167 | 2556 | TPHMARCM-M001533_seg001 | Trh-Gal4 | M | 41582.08 | 1.00 |
| fru-M-700072 | 997 | FruMARCM-M001178_seg001 | fru-Gal4 | M | 25266.43 | 0.66 |
| fru-M-600043 | 1003 | FruMARCM-M001184_seg002 | fru-Gal4 | M | 27702.56 | 0.70 |
| fru-M-100118 | 1126 | FruMARCM-M001322_seg001 | fru-Gal4 | M | 27295.66 | 0.68 |
| fru-M-000069 | 1192 | FruMARCM-M001372_seg001 | fru-Gal4 | M | 28533.90 | 0.69 |
| fru-M-800053 | 1228 | FruMARCM-M001411_seg001 | fru-Gal4 | M | 27644.58 | 0.69 |
| fru-M-500061 | 1367 | FruMARCM-M000877_seg001 | fru-Gal4 | M | 27478.90 | 0.68 |
| fru-M-700063 | 1397 | FruMARCM-M000898_seg001 | fru-Gal4 | M | 27189.54 | 0.68 |
| fru-M-200078 | 1598 | FruMARCM-M000693_seg001 | fru-Gal4 | M | 22629.22 | 0.59 |
| Trh-M-000137 | 1729 | TPHMARCM-M001805_seg001 | Trh-Gal4 | M | 26105.58 | 0.65 |
| Trh-M-000147 | 1739 | TPHMARCM-M001816_seg001 | Trh-Gal4 | M | 26303.66 | 0.67 |
| Trh-M-000155 | 1746 | TPHMARCM-M001841_seg001 | Trh-Gal4 | M | 27278.43 | 0.67 |
| fru-M-100027 | 1773 | FruMARCM-M000290_seg001 | fru-Gal4 | M | 24072.11 | 0.63 |
| fru-M-200044 | 1782 | FruMARCM-M000301_seg001 | fru-Gal4 | M | 23906.05 | 0.61 |
| fru-M-200045 | 1783 | FruMARCM-M000302_seg001 | fru-Gal4 | M | 28854.49 | 0.70 |
| fru-M-300065 | 1923 | FruMARCM-M000518_seg001 | fru-Gal4 | M | 27372.98 | 0.66 |
| Trh-M-300065 | 1983 | TPHMARCM-0688M_seg001 | Trh-Gal4 | M | 27053.58 | 0.68 |
| Trh-M-600036 | 1988 | TPHMARCM-0772M_seg001 | Trh-Gal4 | M | 27900.54 | 0.70 |
| Trh-M-600110 | 2002 | TPHMARCM-M001699_seg001 | Trh-Gal4 | M | 26648.49 | 0.64 |
| Trh-M-600080 | 2041 | TPHMARCM-M001551_seg001 | Trh-Gal4 | M | 24847.66 | 0.64 |
| Trh-M-900044 | 2054 | TPHMARCM-M001559_seg001 | Trh-Gal4 | M | 27436.28 | 0.68 |
| Trh-M-900049 | 2060 | TPHMARCM-M001565_seg001 | Trh-Gal4 | M | 26028.34 | 0.67 |
| Trh-M-500196 | 2125 | TPHMARCM-M001618_seg001 | Trh-Gal4 | M | 28222.91 | 0.65 |
| Trh-M-500197 | 2209 | TPHMARCM-M001700_seg001 | Trh-Gal4 | M | 28208.41 | 0.70 |
| Trh-M-000065 | 2212 | TPHMARCM-M001703_seg001 | Trh-Gal4 | M | 26452.01 | 0.66 |
| Trh-M-000073 | 2242 | TPHMARCM-M001729_seg001 | Trh-Gal4 | M | 27922.21 | 0.69 |
| Trh-M-000074 | 2243 | TPHMARCM-M001730_seg001 | Trh-Gal4 | M | 28329.71 | 0.70 |
| Trh-M-000078 | 2248 | TPHMARCM-M001735_seg001 | Trh-Gal4 | M | 27407.48 | 0.66 |
| Trh-M-000079 | 2249 | TPHMARCM-M001736_seg001 | Trh-Gal4 | M | 28211.63 | 0.71 |
| Trh-M-000083 | 2254 | TPHMARCM-M001741_seg001 | Trh-Gal4 | M | 17914.43 | 0.50 |
| Trh-M-000084 | 2255 | TPHMARCM-M001742_seg001 | Trh-Gal4 | M | 27310.20 | 0.67 |
| Trh-M-000113 | 2299 | TPHMARCM-M001781_seg001 | Trh-Gal4 | M | 28311.06 | 0.70 |
| Trh-M-000117 | 2303 | TPHMARCM-M001785_seg001 | Trh-Gal4 | M | 25771.60 | 0.63 |
| Trh-M-000121 | 2307 | TPHMARCM-M001789_seg001 | Trh-Gal4 | M | 28347.47 | 0.69 |
| Trh-M-000127 | 2313 | TPHMARCM-M001795_seg001 | Trh-Gal4 | M | 28249.63 | 0.68 |
| Trh-M-000026 | 2652 | TPHMARCM-M001421_seg001 | Trh-Gal4 | M | 27858.61 | 0.69 |
| Trh-M-100081 | 2666 | TPHMARCM-M001430_seg001 | Trh-Gal4 | M | 27764.25 | 0.68 |
| Trh-M-100082 | 2667 | TPHMARCM-M001431_seg001 | Trh-Gal4 | M | 29541.51 | 0.71 |
| Trh-M-100083 | 2668 | TPHMARCM-M001432_seg001 | Trh-Gal4 | M | 28242.38 | 0.69 |
| Trh-M-000033 | 2671 | TPHMARCM-M001437_seg001 | Trh-Gal4 | M | 27286.94 | 0.68 |
| Trh-M-500107 | 2695 | TPHMARCM-M001462_seg001 | Trh-Gal4 | M | 29196.50 | 0.70 |
| Trh-M-600075 | 2728 | TPHMARCM-M001492_seg001 | Trh-Gal4 | M | 28398.09 | 0.71 |
| Trh-M-500143 | 2740 | TPHMARCM-M001506_seg001 | Trh-Gal4 | M | 28101.42 | 0.67 |
| Trh-M-500088 | 2767 | TPHMARCM-1114M_seg1 | Trh-Gal4 | M | 29103.23 | 0.67 |
| Trh-M-700053 | 2803 | TPHMARCM-1031M_seg1 | Trh-Gal4 | M | 27117.76 | 0.69 |
| Trh-M-700061 | 2813 | TPHMARCM-1042M_seg1 | Trh-Gal4 | M | 27289.31 | 0.64 |
| Trh-M-400057 | 2816 | TPHMARCM-1045M_seg1 | Trh-Gal4 | M | 26304.17 | 0.65 |
| Trh-M-400058 | 2818 | TPHMARCM-1047M_seg1 | Trh-Gal4 | M | 27185.37 | 0.66 |
| Trh-M-400059 | 2822 | TPHMARCM-1051M_seg1 | Trh-Gal4 | M | 28582.55 | 0.71 |
| Trh-M-100053 | 2921 | TPHMARCM-856M_seg1 | Trh-Gal4 | M | 27061.69 | 0.67 |
| Trh-M-200040 | 2930 | TPHMARCM-865M_seg1 | Trh-Gal4 | M | 26835.89 | 0.65 |
| Trh-M-900008 | 2936 | TPHMARCM-871M_seg1 | Trh-Gal4 | M | 27493.04 | 0.66 |
| Trh-M-900009 | 2937 | TPHMARCM-877M_seg1 | Trh-Gal4 | M | 25553.25 | 0.62 |
| Trh-M-900018 | 2955 | TPHMARCM-896M_seg1 | Trh-Gal4 | M | 27320.22 | 0.67 |
| Trh-M-600053 | 3027 | TPHMARCM-979M_seg1 | Trh-Gal4 | M | 25866.95 | 0.65 |
| Trh-M-700033 | 3041 | TPHMARCM-995M_seg1 | Trh-Gal4 | M | 28453.44 | 0.70 |
| Trh-M-000002 | 3397 | TPHMARCM-083M_seg1 | Trh-Gal4 | M | 27432.90 | 0.67 |
| Trh-M-000003 | 3398 | TPHMARCM-084M_seg1 | Trh-Gal4 | M | 28563.37 | 0.71 |
| Trh-M-000007 | 3403 | TPHMARCM-089M_seg1 | Trh-Gal4 | M | 28877.36 | 0.71 |
| Trh-M-200012 | 3445 | TPHMARCM-194M_seg1 | Trh-Gal4 | M | 26975.38 | 0.68 |
| Trh-M-100032 | 3472 | TPHMARCM-342M_seg1 | Trh-Gal4 | M | 26369.91 | 0.67 |
| Trh-M-200023 | 3485 | TPHMARCM-358M_seg1 | Trh-Gal4 | M | 28162.59 | 0.70 |
| Trh-M-100047 | 3576 | TPHMARCM-506M_seg1 | Trh-Gal4 | M | 29510.62 | 0.69 |
| Trh-M-000021 | 3579 | TPHMARCM-516M_seg1 | Trh-Gal4 | M | 26413.88 | 0.64 |
| Trh-M-500034 | 3622 | TPHMARCM-596M_seg1 | Trh-Gal4 | M | 24147.54 | 0.61 |
| Trh-M-800003 | 3638 | TPHMARCM-628M_seg1 | Trh-Gal4 | M | 27342.55 | 0.69 |
| Trh-M-600033 | 3669 | TPHMARCM-744M_seg1 | Trh-Gal4 | M | 27678.11 | 0.67 |
| Trh-M-500054 | 3674 | TPHMARCM-774M_seg1 | Trh-Gal4 | M | 28304.26 | 0.68 |
| Trh-M-200036 | 3699 | TPHMARCM-850M_seg1 | Trh-Gal4 | M | 26211.09 | 0.68 |
| fru-F-300022 | 9783 | FruMARCM-F000304_seg001 | fru-Gal4 | F | 19631.52 | 0.57 |
| Trh-F-000084 | 10062 | TPHMARCM-F001846_seg001 | Trh-Gal4 | F | 25607.69 | 0.61 |
| Trh-F-100029 | 13838 | TPHMARCM-076F_seg1 | Trh-Gal4 | F | 23678.20 | 0.62 |
| Trh-F-200043 | 13989 | TPHMARCM-278F_seg1 | Trh-Gal4 | F | 28208.92 | 0.69 |
| Trh-F-100051 | 14117 | TPHMARCM-477F_seg1 | Trh-Gal4 | F | 27764.85 | 0.68 |
| Trh-F-500072 | 14235 | TPHMARCM-687F_seg1 | Trh-Gal4 | F | 27050.52 | 0.64 |