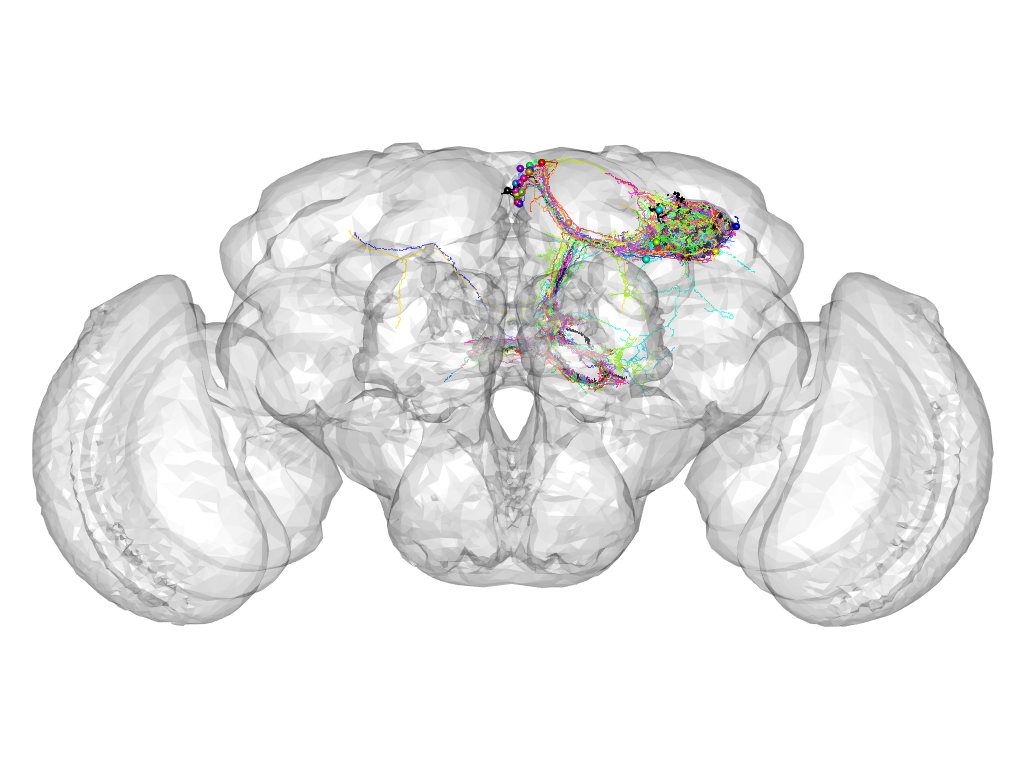
Cluster 825
Part of supercluster XVIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-200355), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 578 | VGlut-F-500688 | XVIII | 0.58 |
| 349 | Cha-F-200259 | XVIII | 0.54 |
| 856 | TH-F-200079 | XVIII | 0.50 |
| 364 | Cha-F-600143 | XVIII | 0.48 |
| 1025 | VGlut-F-000057 | XVIII | 0.35 |
Cluster composition
This cluster has 22 neurons. The exemplar of this cluster (VGlut-F-200355) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-200355 | 12972 | DvGlutMARCM-F2007_seg2 | VGlut-Gal4 | F | 57527.00 | 1.00 |
| fru-M-200270 | 91 | FruMARCM-M002290_seg001 | fru-Gal4 | M | 43423.46 | 0.77 |
| fru-M-200288 | 162 | FruMARCM-M002382_seg003 | fru-Gal4 | M | 42460.63 | 0.74 |
| fru-M-700172 | 203 | FruMARCM-M002418_seg001 | fru-Gal4 | M | 42262.06 | 0.73 |
| fru-M-300222 | 709 | FruMARCM-M001912_seg001 | fru-Gal4 | M | 34453.59 | 0.58 |
| fru-M-600092 | 885 | FruMARCM-M002148_seg002 | fru-Gal4 | M | 42112.76 | 0.71 |
| fru-M-500082 | 1405 | FruMARCM-M000904_seg001 | fru-Gal4 | M | 39555.21 | 0.60 |
| fru-M-100024 | 1770 | FruMARCM-M000288_seg001 | fru-Gal4 | M | 41434.73 | 0.75 |
| Cha-F-000298 | 4190 | ChaMARCM-F001292_seg001 | Cha-Gal4 | F | 36430.50 | 0.71 |
| fru-F-100053 | 4508 | FruMARCM-F001618_seg001 | fru-Gal4 | F | 41593.34 | 0.73 |
| fru-F-500213 | 4600 | FruMARCM-F001719_seg001 | fru-Gal4 | F | 43580.80 | 0.77 |
| Cha-F-600160 | 5122 | ChaMARCM-F001177_seg002 | Cha-Gal4 | F | 30283.55 | 0.57 |
| Cha-F-300159 | 7138 | ChaMARCM-F000605_seg001 | Cha-Gal4 | F | 36787.59 | 0.59 |
| Gad1-F-500086 | 9703 | GadMARCM-F000547_seg001 | Gad1-Gal4 | F | 43786.86 | 0.76 |
| fru-F-100027 | 10024 | FruMARCM-F000608_seg002 | fru-Gal4 | F | 42302.21 | 0.77 |
| fru-F-600000 | 11450 | FruMARCM-F000067_seg001 | fru-Gal4 | F | 32070.95 | 0.53 |
| VGlut-F-000453 | 11796 | DvGlutMARCM-F002361_seg001 | VGlut-Gal4 | F | 42322.59 | 0.76 |
| VGlut-F-000465 | 11933 | DvGlutMARCM-F002468_seg001 | VGlut-Gal4 | F | 43152.23 | 0.77 |
| VGlut-F-400325 | 14805 | DvGlutMARCM-F1152_seg1 | VGlut-Gal4 | F | 43483.25 | 0.75 |
| VGlut-F-000354 | 15052 | DvGlutMARCM-F1391_seg1 | VGlut-Gal4 | F | 42775.56 | 0.76 |
| VGlut-F-200275 | 15071 | DvGlutMARCM-F1409_seg1 | VGlut-Gal4 | F | 42869.02 | 0.74 |
| VGlut-F-200133 | 15524 | DvGlutMARCM-F671_seg1 | VGlut-Gal4 | F | 43662.38 | 0.76 |