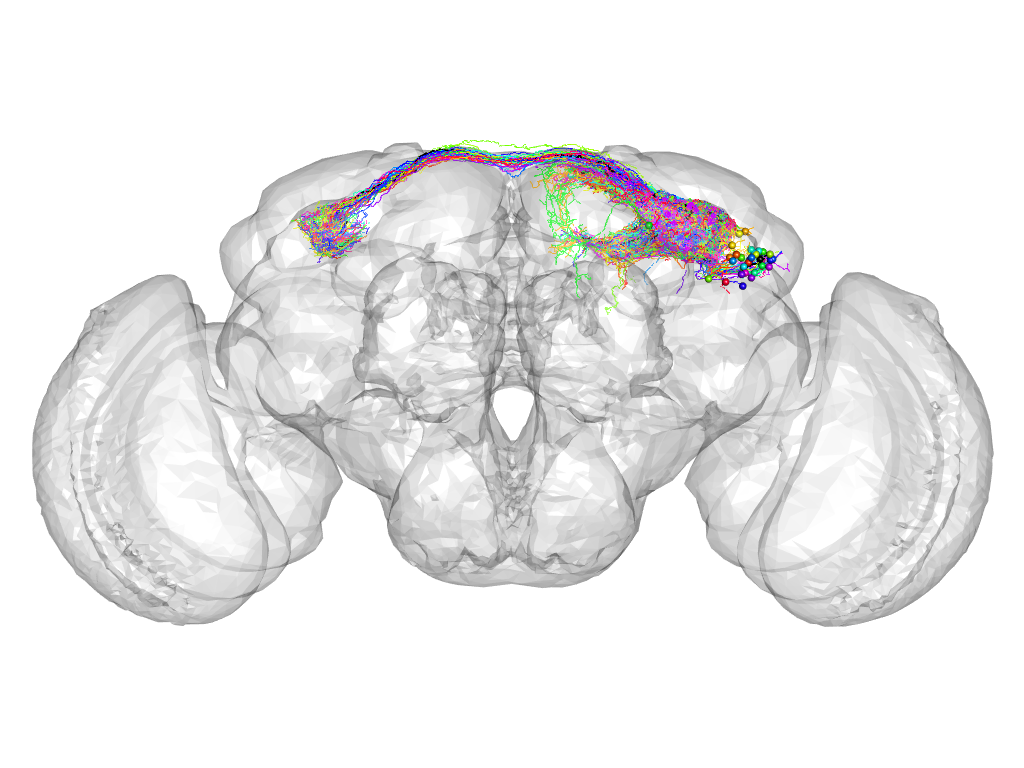
Cluster 578
Part of supercluster XVIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-500688), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 856 | TH-F-200079 | XVIII | 0.62 |
| 825 | VGlut-F-200355 | XVIII | 0.57 |
| 349 | Cha-F-200259 | XVIII | 0.53 |
| 1025 | VGlut-F-000057 | XVIII | 0.50 |
| 203 | Trh-M-000010 | XVIII | 0.44 |
Cluster composition
This cluster has 36 neurons. The exemplar of this cluster (VGlut-F-500688) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-500688 | 8951 | DvGlutMARCM-F003515_seg001 | VGlut-Gal4 | F | 49520.37 | 1.00 |
| fru-M-200271 | 92 | FruMARCM-M002291_seg001 | fru-Gal4 | M | 38390.80 | 0.77 |
| fru-M-900058 | 112 | FruMARCM-M002309_seg002 | fru-Gal4 | M | 38443.16 | 0.77 |
| fru-M-700181 | 283 | FruMARCM-M002509_seg001 | fru-Gal4 | M | 38750.00 | 0.77 |
| fru-M-200304 | 297 | FruMARCM-M002525_seg001 | fru-Gal4 | M | 38039.67 | 0.76 |
| fru-M-200314 | 317 | FruMARCM-M002539_seg003 | fru-Gal4 | M | 28574.90 | 0.58 |
| fru-M-000087 | 556 | FruMARCM-M001728_seg002 | fru-Gal4 | M | 37676.00 | 0.76 |
| fru-M-100173 | 702 | FruMARCM-M001904_seg001 | fru-Gal4 | M | 38184.16 | 0.77 |
| fru-M-200101 | 1480 | FruMARCM-M001037_seg001 | fru-Gal4 | M | 37760.24 | 0.77 |
| fru-M-100082 | 1520 | FruMARCM-M001064_seg003 | fru-Gal4 | M | 37229.19 | 0.76 |
| fru-M-800003 | 2375 | FruMARCM-M000105_seg002 | fru-Gal4 | M | 38609.09 | 0.78 |
| fru-M-800011 | 2481 | FruMARCM-M000236_seg001 | fru-Gal4 | M | 38264.33 | 0.78 |
| fru-F-000118 | 3723 | FruMARCM-F002221_seg001 | fru-Gal4 | F | 39630.36 | 0.79 |
| Gad1-F-000087 | 4095 | GadMARCM-F000685_seg001 | Gad1-Gal4 | F | 30219.83 | 0.54 |
| fru-F-000110 | 4773 | FruMARCM-F002091_seg001 | fru-Gal4 | F | 39459.04 | 0.78 |
| fru-F-000069 | 6400 | FruMARCM-F001535_seg001 | fru-Gal4 | F | 39137.80 | 0.79 |
| VGlut-F-600780 | 6452 | DvGlutMARCM-F004236_seg001 | VGlut-Gal4 | F | 37915.31 | 0.77 |
| VGlut-F-500739 | 6712 | DvGlutMARCM-F003924_seg001 | VGlut-Gal4 | F | 38077.88 | 0.77 |
| VGlut-F-000636 | 6950 | DvGlutMARCM-F004104_seg001 | VGlut-Gal4 | F | 37118.37 | 0.76 |
| VGlut-F-000498 | 9275 | DvGlutMARCM-F003330_seg001 | VGlut-Gal4 | F | 39366.33 | 0.79 |
| VGlut-F-500592 | 9357 | DvGlutMARCM-F003189_seg001 | VGlut-Gal4 | F | 38236.64 | 0.78 |
| VGlut-F-500489 | 11016 | DvGlutMARCM-F002899_seg001 | VGlut-Gal4 | F | 37843.11 | 0.77 |
| VGlut-F-000427 | 12943 | DvGlutMARCM-F1977_seg2 | VGlut-Gal4 | F | 38049.29 | 0.77 |
| VGlut-F-000198 | 14564 | DvGlutMARCM-F828_seg1 | VGlut-Gal4 | F | 38601.95 | 0.78 |
| VGlut-F-100108 | 14782 | DvGlutMARCM-F1129_seg1 | VGlut-Gal4 | F | 38897.53 | 0.78 |
| VGlut-F-100131 | 14912 | DvGlutMARCM-F1255_seg1 | VGlut-Gal4 | F | 33500.04 | 0.72 |
| VGlut-F-100151 | 15072 | DvGlutMARCM-F1410_seg1 | VGlut-Gal4 | F | 37365.78 | 0.76 |
| VGlut-F-000400 | 15389 | DvGlutMARCM-F1747_seg1 | VGlut-Gal4 | F | 37725.27 | 0.77 |
| VGlut-F-500003 | 15597 | DvGlutMARCM-F013_seg1 | VGlut-Gal4 | F | 38042.28 | 0.77 |
| VGlut-F-200030 | 15639 | DvGlutMARCM-F053_seg1 | VGlut-Gal4 | F | 39071.68 | 0.77 |
| VGlut-F-000029 | 15789 | DvGlutMARCM-F200_seg1 | VGlut-Gal4 | F | 37014.03 | 0.76 |
| VGlut-F-300042 | 15918 | DvGlutMARCM-F313_seg1 | VGlut-Gal4 | F | 37363.88 | 0.76 |
| VGlut-F-100036 | 16043 | DvGlutMARCM-F434_seg1 | VGlut-Gal4 | F | 39733.32 | 0.79 |
| VGlut-F-000131 | 16142 | DvGlutMARCM-F525_seg1 | VGlut-Gal4 | F | 37131.19 | 0.76 |
| VGlut-F-000132 | 16143 | DvGlutMARCM-F526_seg1 | VGlut-Gal4 | F | 38333.37 | 0.77 |
| VGlut-F-000147 | 16167 | DvGlutMARCM-F548_seg2 | VGlut-Gal4 | F | 36071.15 | 0.73 |