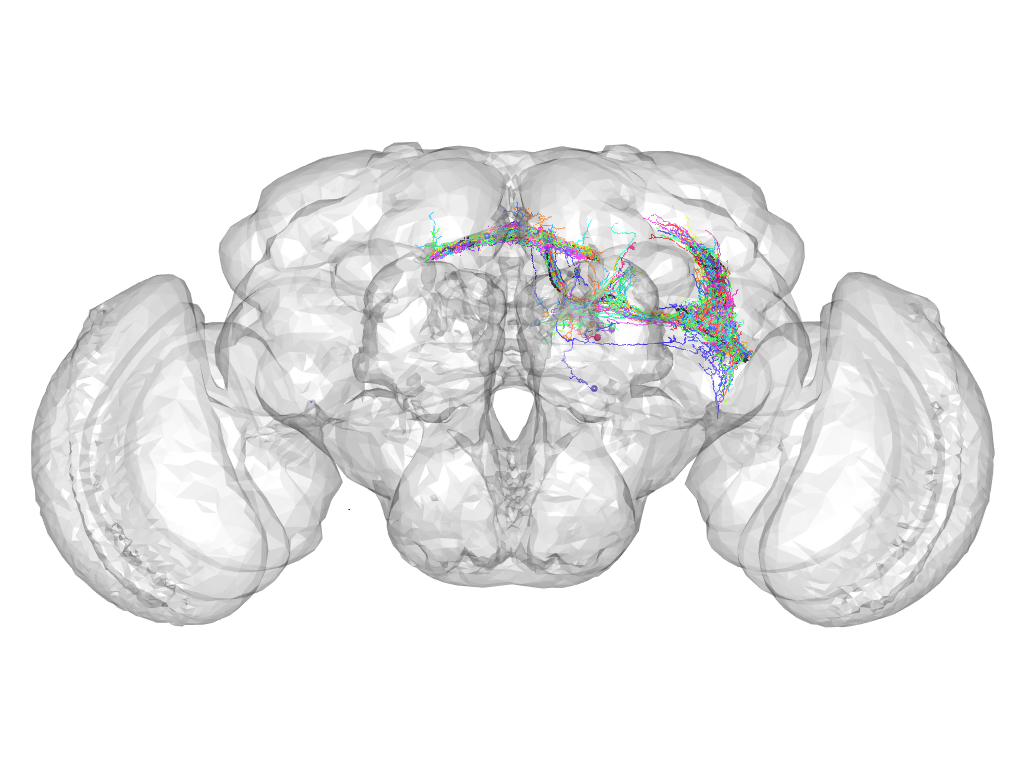
Cluster 808
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-300136), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 875 | Trh-F-100018 | XIX | 0.72 |
| 964 | VGlut-F-200325 | XIV | 0.34 |
| 461 | Cha-F-900029 | XVII | 0.31 |
| 904 | VGlut-F-100092 | XVII | 0.28 |
| 621 | Gad1-F-500089 | XVII | 0.26 |
Cluster composition
This cluster has 29 neurons. The exemplar of this cluster (Trh-F-300136) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-300136 | 12704 | TPHMARCM-1155F_seg1 | Trh-Gal4 | F | 13097.61 | 1.00 |
| Trh-M-400120 | 2104 | TPHMARCM-M001603_seg001 | Trh-Gal4 | M | 9648.83 | 0.72 |
| Trh-M-300127 | 2602 | TPHMARCM-1220M_seg1 | Trh-Gal4 | M | 9713.57 | 0.73 |
| Trh-M-200061 | 2630 | TPHMARCM-M001368_seg001 | Trh-Gal4 | M | 8874.12 | 0.62 |
| Trh-M-200062 | 2631 | TPHMARCM-M001369_seg001 | Trh-Gal4 | M | 9417.94 | 0.72 |
| Trh-M-200047 | 2861 | TPHMARCM-1093M_seg1 | Trh-Gal4 | M | 9636.32 | 0.72 |
| Trh-M-100059 | 2874 | TPHMARCM-1118M_seg1 | Trh-Gal4 | M | 9697.22 | 0.72 |
| Trh-M-100065 | 2898 | TPHMARCM-1137M_seg1 | Trh-Gal4 | M | 9469.00 | 0.72 |
| Trh-M-300121 | 2914 | TPHMARCM-1160M_seg1 | Trh-Gal4 | M | 9474.54 | 0.69 |
| Trh-M-300122 | 2915 | TPHMARCM-1161M_seg1 | Trh-Gal4 | M | 10144.78 | 0.75 |
| Trh-M-100023 | 3412 | TPHMARCM-097M_seg1 | Trh-Gal4 | M | 9852.85 | 0.71 |
| Trh-M-300002 | 3439 | TPHMARCM-186M_seg1 | Trh-Gal4 | M | 9815.29 | 0.73 |
| Trh-M-200016 | 3458 | TPHMARCM-319M_seg1 | Trh-Gal4 | M | 10119.55 | 0.72 |
| Trh-M-300010 | 3467 | TPHMARCM-337M_seg1 | Trh-Gal4 | M | 9413.28 | 0.70 |
| Trh-M-300038 | 3535 | TPHMARCM-436M_seg1 | Trh-Gal4 | M | 9537.33 | 0.69 |
| Trh-M-200026 | 3548 | TPHMARCM-471M_seg1 | Trh-Gal4 | M | 9391.96 | 0.71 |
| Trh-M-200030 | 3644 | TPHMARCM-659M_seg1 | Trh-Gal4 | M | 9755.33 | 0.70 |
| Trh-F-200107 | 12699 | TPHMARCM-1148F_seg1 | Trh-Gal4 | F | 9989.97 | 0.74 |
| Trh-F-200091 | 13278 | TPHMARCM-764F_seg1 | Trh-Gal4 | F | 9769.89 | 0.72 |
| Trh-F-100021 | 13814 | TPHMARCM-033F_seg1 | Trh-Gal4 | F | 10061.34 | 0.71 |
| Trh-F-300008 | 13887 | TPHMARCM-152F_seg1 | Trh-Gal4 | F | 4978.96 | 0.34 |
| Trh-F-200028 | 13956 | TPHMARCM-247F-0812-02_seg1 | Trh-Gal4 | F | 9968.36 | 0.76 |
| Trh-F-300024 | 13983 | TPHMARCM-273F_seg1 | Trh-Gal4 | F | 9787.82 | 0.73 |
| Trh-F-200058 | 14015 | TPHMARCM-302F_seg1 | Trh-Gal4 | F | 9871.42 | 0.73 |
| Trh-F-300040 | 14047 | TPHMARCM-375F_seg1 | Trh-Gal4 | F | 9210.77 | 0.71 |
| Trh-F-300046 | 14062 | TPHMARCM-390F_seg1 | Trh-Gal4 | F | 9651.99 | 0.72 |
| Trh-F-200086 | 14232 | TPHMARCM-684F_seg1 | Trh-Gal4 | F | 9847.90 | 0.75 |
| Trh-F-300100 | 14248 | TPHMARCM-699F_seg1 | Trh-Gal4 | F | 9579.63 | 0.69 |
| Trh-F-300117 | 14288 | TPHMARCM-831F_seg1 | Trh-Gal4 | F | 9690.55 | 0.73 |