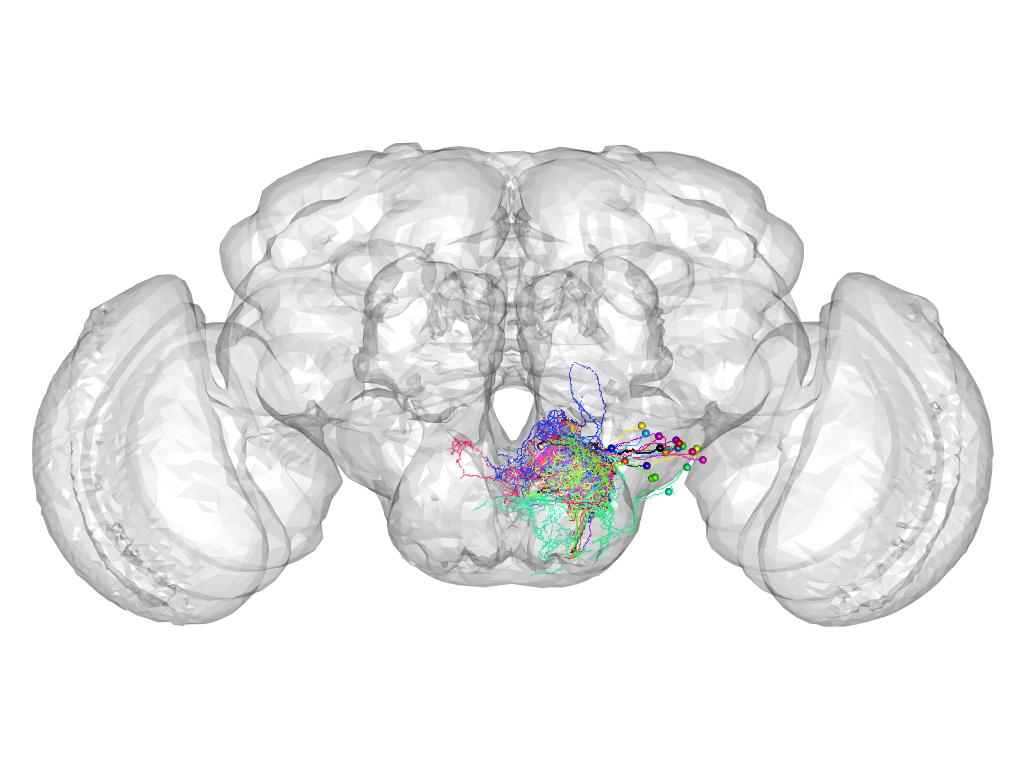
Cluster 797
Part of supercluster IX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-F-200118), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 791 | VGlut-F-000482 | IX | 0.76 |
| 687 | VGlut-F-500173 | IX | 0.70 |
| 863 | TH-F-000022 | XII | 0.61 |
| 810 | VGlut-F-000431 | IX | 0.56 |
| 217 | fru-F-100078 | IX | 0.53 |
Cluster composition
This cluster has 21 neurons. The exemplar of this cluster (Trh-F-200118) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-F-200118 | 12402 | TPHMARCM-1198F_seg1 | Trh-Gal4 | F | 15364.07 | 1.00 |
| Trh-M-400133 | 2150 | TPHMARCM-M001638_seg002 | Trh-Gal4 | M | 11501.48 | 0.75 |
| Trh-M-100098 | 2160 | TPHMARCM-M001649_seg002 | Trh-Gal4 | M | 11081.34 | 0.75 |
| Trh-M-400138 | 2163 | TPHMARCM-M001651_seg002 | Trh-Gal4 | M | 10402.90 | 0.71 |
| Trh-M-300094 | 2850 | TPHMARCM-1078M_seg1 | Trh-Gal4 | M | 11291.76 | 0.73 |
| Trh-M-300119 | 2912 | TPHMARCM-1158M_seg1 | Trh-Gal4 | M | 10472.96 | 0.68 |
| Trh-M-300027 | 3524 | TPHMARCM-412M_seg1 | Trh-Gal4 | M | 10928.44 | 0.72 |
| Trh-M-300043 | 3556 | TPHMARCM-487M_seg1 | Trh-Gal4 | M | 11024.09 | 0.71 |
| Trh-M-300063 | 3659 | TPHMARCM-673M_seg2 | Trh-Gal4 | M | 10726.75 | 0.71 |
| Cha-F-300115 | 7446 | ChaMARCM-F000330_seg002 | Cha-Gal4 | F | 11058.84 | 0.67 |
| Cha-F-800004 | 7491 | ChaMARCM-F000357_seg001 | Cha-Gal4 | F | 10717.46 | 0.51 |
| Trh-F-500182 | 12540 | TPHMARCM-1328F_seg1 | Trh-Gal4 | F | 9726.54 | 0.67 |
| Trh-F-500216 | 12588 | TPHMARCM-F001381_seg002 | Trh-Gal4 | F | 10982.16 | 0.75 |
| Trh-F-400085 | 12722 | TPHMARCM-883F_seg2 | Trh-Gal4 | F | 10946.31 | 0.72 |
| VGlut-F-600138 | 13136 | DvGlutMARCM-F2166_seg1 | VGlut-Gal4 | F | 8377.26 | 0.51 |
| Trh-F-300003 | 13874 | TPHMARCM-143F_seg1 | Trh-Gal4 | F | 10296.01 | 0.71 |
| Trh-F-300005 | 13876 | TPHMARCM-145F_seg1 | Trh-Gal4 | F | 11403.58 | 0.74 |
| Trh-F-300088 | 14184 | TPHMARCM-623F_seg1 | Trh-Gal4 | F | 11463.87 | 0.74 |
| Trh-F-300101 | 14249 | TPHMARCM-699F_seg2 | Trh-Gal4 | F | 10915.19 | 0.71 |
| Trh-F-300116 | 14286 | TPHMARCM-829F_seg1 | Trh-Gal4 | F | 11133.63 | 0.73 |
| VGlut-F-000367 | 15188 | DvGlutMARCM-F1517_seg2 | VGlut-Gal4 | F | 10440.46 | 0.64 |