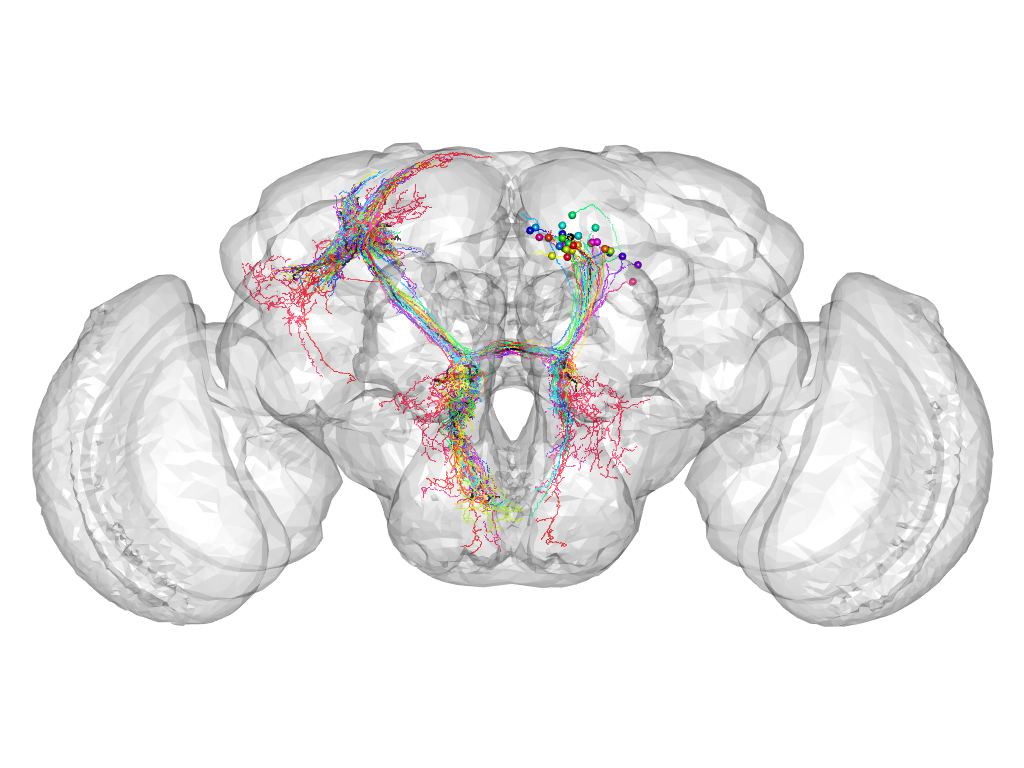
Cluster 74
Part of supercluster XIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-000152), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 307 | fru-F-500216 | XIII | 0.51 |
| 909 | VGlut-F-000297 | XIII | 0.45 |
| 662 | Tdc2-F-400026 | XIII | 0.14 |
| 91 | fru-M-200161 | IX | 0.08 |
| 877 | Trh-F-000012 | XIX | -0.01 |
Cluster composition
This cluster has 30 neurons. The exemplar of this cluster (fru-M-000152) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-000152 | 846 | FruMARCM-M002099_seg001 | fru-Gal4 | M | 20386.72 | 1.00 |
| fru-M-400217 | 230 | FruMARCM-M002443_seg001 | fru-Gal4 | M | 13163.06 | 0.50 |
| fru-M-500247 | 374 | FruMARCM-M002609_seg001 | fru-Gal4 | M | 15697.26 | 0.75 |
| fru-M-600111 | 378 | FruMARCM-M002613_seg001 | fru-Gal4 | M | 14539.35 | 0.72 |
| fru-M-600112 | 392 | FruMARCM-M002624_seg001 | fru-Gal4 | M | 13325.73 | 0.71 |
| fru-M-000083 | 520 | FruMARCM-M001632_seg002 | fru-Gal4 | M | 14143.07 | 0.70 |
| fru-M-700096 | 544 | FruMARCM-M001680_seg001 | fru-Gal4 | M | 12798.51 | 0.66 |
| fru-M-500159 | 553 | FruMARCM-M001715_seg001 | fru-Gal4 | M | 14088.83 | 0.66 |
| fru-M-000117 | 643 | FruMARCM-M001851_seg001 | fru-Gal4 | M | 14435.58 | 0.72 |
| fru-M-500185 | 719 | FruMARCM-M001927_seg001 | fru-Gal4 | M | 11782.81 | 0.68 |
| fru-M-500201 | 781 | FruMARCM-M001990_seg001 | fru-Gal4 | M | 12361.27 | 0.63 |
| fru-M-600091 | 884 | FruMARCM-M002148_seg001 | fru-Gal4 | M | 11739.32 | 0.65 |
| fru-M-700138 | 917 | FruMARCM-M002178_seg001 | fru-Gal4 | M | 12399.46 | 0.68 |
| fru-M-500101 | 977 | FruMARCM-M001151_seg001 | fru-Gal4 | M | 12735.78 | 0.66 |
| fru-M-500110 | 1022 | FruMARCM-M001204_seg002 | fru-Gal4 | M | 15118.17 | 0.70 |
| fru-M-400133 | 1121 | FruMARCM-M001318_seg002 | fru-Gal4 | M | 9535.53 | 0.48 |
| fru-M-100133 | 1175 | FruMARCM-M001358_seg003 | fru-Gal4 | M | 13809.63 | 0.70 |
| fru-M-400140 | 1237 | FruMARCM-M001419_seg001 | fru-Gal4 | M | 14279.46 | 0.67 |
| fru-M-600063 | 1266 | FruMARCM-M001456_seg001 | fru-Gal4 | M | 11863.95 | 0.64 |
| fru-M-500144 | 1311 | FruMARCM-M001529_seg001 | fru-Gal4 | M | 14393.63 | 0.68 |
| fru-M-400102 | 1338 | FruMARCM-M001093_seg002 | fru-Gal4 | M | 13555.80 | 0.65 |
| fru-M-500098 | 1557 | FruMARCM-M001089_seg001 | fru-Gal4 | M | 13473.29 | 0.70 |
| fru-M-600016 | 1759 | FruMARCM-M000249_seg001 | fru-Gal4 | M | 13248.10 | 0.69 |
| fru-M-700031 | 1870 | FruMARCM-M000413_seg002 | fru-Gal4 | M | 11419.57 | 0.61 |
| fru-M-700040 | 1896 | FruMARCM-M000427_seg001 | fru-Gal4 | M | 13465.11 | 0.70 |
| fru-M-500022 | 1914 | FruMARCM-M000469_seg001 | fru-Gal4 | M | 13671.06 | 0.69 |
| fru-M-500043 | 1969 | FruMARCM-M000616_seg001 | fru-Gal4 | M | 14971.25 | 0.74 |
| fru-M-300005 | 2346 | FruMARCM-M000038_seg001 | fru-Gal4 | M | 12641.30 | 0.69 |
| 5-HT1B-M-500005 | 2486 | 5HT1bMARCM-M000040_seg001 | 5-HT1B-Gal4 | M | 14174.71 | 0.57 |
| VGlut-F-400486 | 13014 | DvGlutMARCM-F2040_seg1 | VGlut-Gal4 | F | 10327.58 | 0.31 |