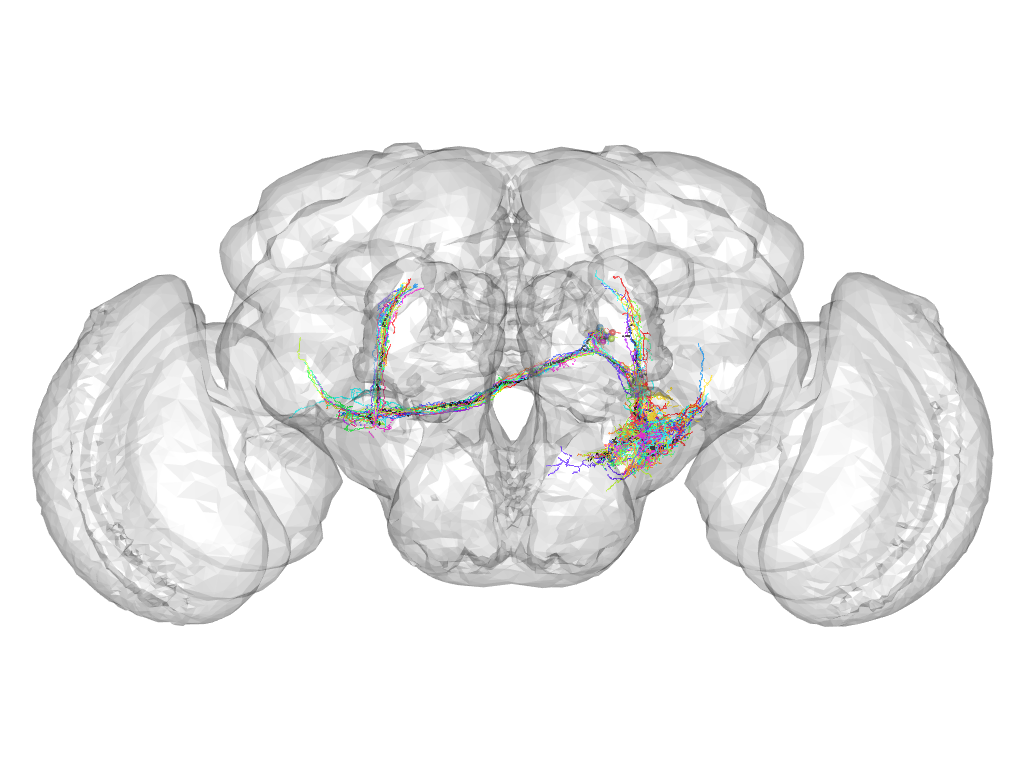
Cluster 637
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-700051), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 652 | Tdc2-F-200010 | VIII | 0.36 |
| 988 | VGlut-F-200005 | X | 0.32 |
| 902 | VGlut-F-400300 | VIII | 0.31 |
| 410 | fru-F-400147 | VIII | 0.30 |
| 855 | TH-F-200057 | VIII | 0.30 |
Cluster composition
This cluster has 15 neurons. The exemplar of this cluster (fru-F-700051) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-700051 | 9971 | FruMARCM-F000562_seg001 | fru-Gal4 | F | 17630.53 | 1.00 |
| fru-M-400215 | 146 | FruMARCM-M002373_seg001 | fru-Gal4 | M | 11300.64 | 0.62 |
| fru-M-500234 | 198 | FruMARCM-M002413_seg001 | fru-Gal4 | M | 12927.26 | 0.68 |
| fru-M-700014 | 953 | FruMARCM-M000151_seg005 | fru-Gal4 | M | 12154.93 | 0.64 |
| fru-M-500065 | 1371 | FruMARCM-M000880_seg001 | fru-Gal4 | M | 12837.39 | 0.65 |
| fru-M-800028 | 1661 | FruMARCM-M000810_seg001 | fru-Gal4 | M | 12476.10 | 0.71 |
| Trh-M-500184 | 2089 | TPHMARCM-M001590_seg001 | Trh-Gal4 | M | 10703.53 | 0.64 |
| fru-M-300027 | 2458 | FruMARCM-M000217_seg001 | fru-Gal4 | M | 12133.84 | 0.67 |
| fru-F-500269 | 3737 | FruMARCM-F002331_seg003 | fru-Gal4 | F | 13120.93 | 0.65 |
| fru-F-400154 | 6359 | FruMARCM-F001502_seg003 | fru-Gal4 | F | 9975.64 | 0.57 |
| VGlut-F-800253 | 6953 | DvGlutMARCM-F004107_seg001 | VGlut-Gal4 | F | 12847.33 | 0.71 |
| Cha-F-300122 | 7222 | ChaMARCM-F000389_seg003 | Cha-Gal4 | F | 10914.66 | 0.59 |
| fru-F-400087 | 7851 | FruMARCM-F000761_seg001 | fru-Gal4 | F | 12637.68 | 0.71 |
| VGlut-F-700424 | 8015 | DvGlutMARCM-F003831_seg002 | VGlut-Gal4 | F | 12909.16 | 0.73 |
| Gad1-F-200050 | 10693 | GadMARCM-F000193_seg001 | Gad1-Gal4 | F | 12401.91 | 0.70 |