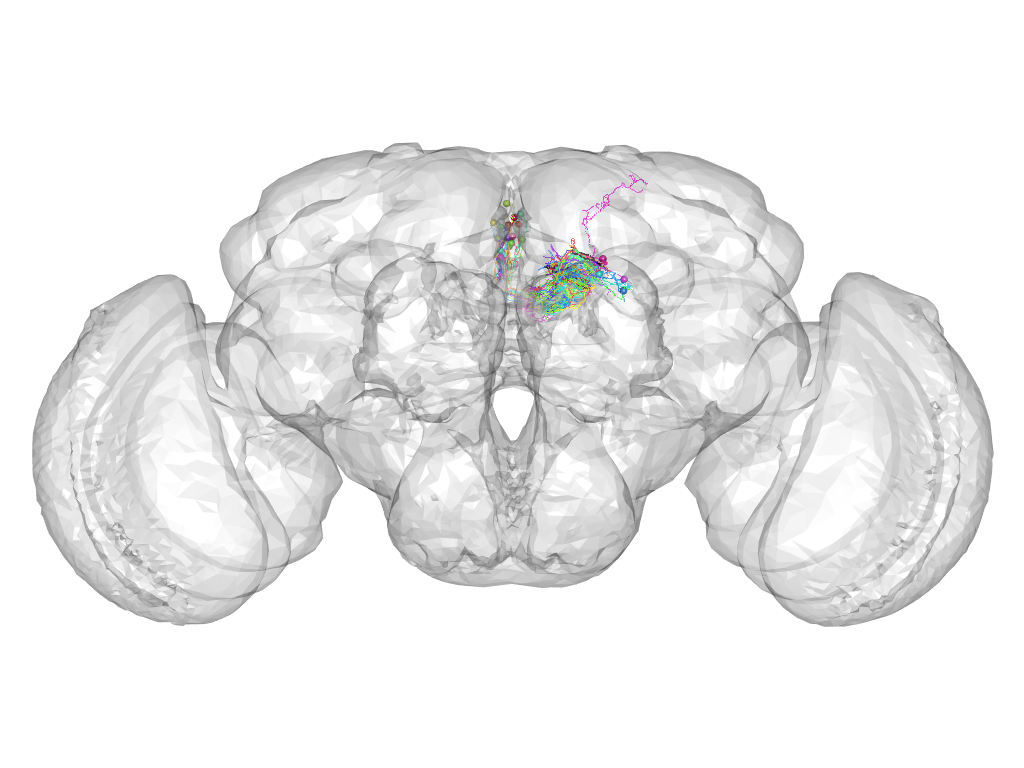
Cluster 299
Part of supercluster XIV
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-400172), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 1028 | VGlut-F-000067 | XIV | 0.61 |
| 84 | fru-M-200026 | XIV | 0.60 |
| 204 | Trh-M-100025 | XIV | 0.59 |
| 631 | fru-F-500016 | XIV | 0.56 |
| 964 | VGlut-F-200325 | XIV | 0.55 |
| 703 | VGlut-F-300427 | XIV | 0.53 |
| 265 | Cha-F-100295 | XIV | 0.52 |
| 192 | TH-M-300027 | XIV | 0.51 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (fru-F-400172) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-400172 | 4537 | FruMARCM-F001644_seg003 | fru-Gal4 | F | 6890.48 | 1.00 |
| fru-M-700163 | 182 | FruMARCM-M002399_seg001 | fru-Gal4 | M | 5202.62 | 0.73 |
| fru-M-500161 | 568 | FruMARCM-M001754_seg001 | fru-Gal4 | M | 5146.61 | 0.73 |
| fru-M-400112 | 974 | FruMARCM-M001147_seg001 | fru-Gal4 | M | 4479.20 | 0.68 |
| fru-M-500130 | 1075 | FruMARCM-M001251_seg001 | fru-Gal4 | M | 5316.77 | 0.77 |
| fru-M-200189 | 1213 | FruMARCM-M001398_seg001 | fru-Gal4 | M | 4520.67 | 0.60 |
| fru-M-200095 | 1446 | FruMARCM-M001015_seg001 | fru-Gal4 | M | 4966.34 | 0.70 |
| fru-M-200097 | 1450 | FruMARCM-M001018_seg001 | fru-Gal4 | M | 4863.69 | 0.66 |
| fru-M-800034 | 1701 | FruMARCM-M000854_seg001 | fru-Gal4 | M | 4842.85 | 0.66 |
| fru-M-600004 | 2412 | FruMARCM-M000133_seg001 | fru-Gal4 | M | 4321.47 | 0.62 |
| Trh-M-400042 | 2972 | TPHMARCM-912M_seg1 | Trh-Gal4 | M | 4896.14 | 0.67 |
| fru-F-500261 | 3713 | FruMARCM-F002207_seg001 | fru-Gal4 | F | 5142.47 | 0.71 |
| fru-F-500267 | 3735 | FruMARCM-F002331_seg001 | fru-Gal4 | F | 5191.40 | 0.72 |
| fru-F-300094 | 4540 | FruMARCM-F001653_seg001 | fru-Gal4 | F | 5339.96 | 0.74 |
| fru-F-300108 | 4647 | FruMARCM-F001813_seg002 | fru-Gal4 | F | 5134.43 | 0.72 |
| Cha-F-500151 | 5545 | ChaMARCM-F000847_seg001 | Cha-Gal4 | F | 4116.99 | 0.56 |
| fru-F-600062 | 6223 | FruMARCM-F001159_seg001 | fru-Gal4 | F | 5121.43 | 0.71 |
| fru-F-500188 | 6373 | FruMARCM-F001511_seg001 | fru-Gal4 | F | 4578.02 | 0.60 |
| fru-F-700097 | 7342 | FruMARCM-F000978_seg001 | fru-Gal4 | F | 4955.03 | 0.71 |
| fru-F-500014 | 9846 | FruMARCM-F000431_seg001 | fru-Gal4 | F | 5202.45 | 0.73 |
| fru-F-700041 | 9901 | FruMARCM-F000490_seg002 | fru-Gal4 | F | 5086.56 | 0.72 |
| Trh-F-000063 | 10043 | TPHMARCM-F001825_seg001 | Trh-Gal4 | F | 5098.04 | 0.72 |
| Trh-F-000066 | 10046 | TPHMARCM-F001828_seg001 | Trh-Gal4 | F | 5160.94 | 0.70 |
| Cha-F-500027 | 10169 | ChaMARCM-F000099_seg002 | Cha-Gal4 | F | 4888.05 | 0.71 |
| Gad1-F-900019 | 11215 | GadMARCM-F000200_seg002 | Gad1-Gal4 | F | 4416.62 | 0.43 |
| Gad1-F-100018 | 11260 | GadMARCM-F000238_seg002 | Gad1-Gal4 | F | 5173.96 | 0.76 |
| Trh-F-700045 | 12715 | TPHMARCM-876F_seg1 | Trh-Gal4 | F | 5186.34 | 0.73 |
| Trh-F-400033 | 13969 | TPHMARCM-259F_seg1 | Trh-Gal4 | F | 4788.24 | 0.65 |