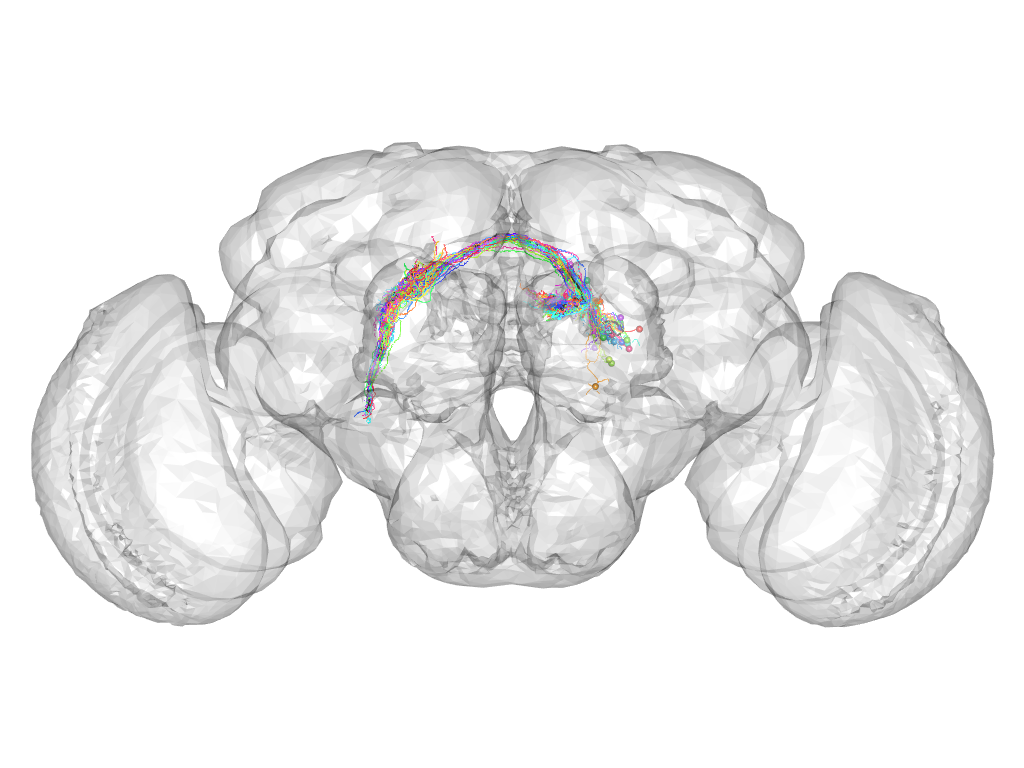
Cluster 325
Part of supercluster XIV
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Cha-F-100151), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 265 | Cha-F-100295 | XIV | 0.63 |
| 245 | Gad1-F-200134 | XIV | 0.49 |
| 964 | VGlut-F-200325 | XIV | 0.43 |
| 351 | Cha-F-100231 | XIV | 0.38 |
| 840 | Tdc2-F-100009 | XIV | 0.37 |
Cluster composition
This cluster has 33 neurons. The exemplar of this cluster (Cha-F-100151) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Cha-F-100151 | 4896 | ChaMARCM-F001012_seg001 | Cha-Gal4 | F | 13541.79 | 1.00 |
| Gad1-F-300134 | 3943 | GadMARCM-F000573_seg001 | Gad1-Gal4 | F | 10427.85 | 0.76 |
| Gad1-F-800076 | 4016 | GadMARCM-F000628_seg001 | Gad1-Gal4 | F | 10529.14 | 0.78 |
| Gad1-F-000084 | 4091 | GadMARCM-F000680_seg001 | Gad1-Gal4 | F | 10476.14 | 0.77 |
| Cha-F-100333 | 4386 | ChaMARCM-F001439_seg001 | Cha-Gal4 | F | 9646.88 | 0.71 |
| Cha-F-000162 | 4848 | ChaMARCM-F000971_seg001 | Cha-Gal4 | F | 10057.63 | 0.76 |
| Cha-F-200206 | 4970 | ChaMARCM-F001067_seg001 | Cha-Gal4 | F | 10507.71 | 0.78 |
| Cha-F-400150 | 5288 | ChaMARCM-F000678_seg001 | Cha-Gal4 | F | 10705.52 | 0.79 |
| Cha-F-200136 | 5310 | ChaMARCM-F000692_seg001 | Cha-Gal4 | F | 9859.87 | 0.72 |
| Cha-F-400157 | 5330 | ChaMARCM-F000706_seg001 | Cha-Gal4 | F | 9790.89 | 0.73 |
| Cha-F-800027 | 5376 | ChaMARCM-F000741_seg001 | Cha-Gal4 | F | 10319.83 | 0.76 |
| Cha-F-600101 | 5413 | ChaMARCM-F000766_seg001 | Cha-Gal4 | F | 10167.14 | 0.77 |
| Cha-F-300229 | 5498 | ChaMARCM-F000818_seg001 | Cha-Gal4 | F | 9868.99 | 0.74 |
| Cha-F-200180 | 5636 | ChaMARCM-F000914_seg001 | Cha-Gal4 | F | 11086.96 | 0.81 |
| Cha-F-200183 | 5642 | ChaMARCM-F000919_seg001 | Cha-Gal4 | F | 9975.50 | 0.75 |
| Gad1-F-300057 | 6012 | GadMARCM-F000205_seg001 | Gad1-Gal4 | F | 10676.55 | 0.80 |
| Cha-F-900028 | 7103 | ChaMARCM-F000578_seg001 | Cha-Gal4 | F | 10034.68 | 0.72 |
| Cha-F-000098 | 7134 | ChaMARCM-F000602_seg001 | Cha-Gal4 | F | 9452.49 | 0.70 |
| Cha-F-300170 | 7175 | ChaMARCM-F000628_seg001 | Cha-Gal4 | F | 9982.00 | 0.72 |
| Cha-F-300172 | 7203 | ChaMARCM-F000647_seg001 | Cha-Gal4 | F | 10407.59 | 0.77 |
| Cha-F-600028 | 7454 | ChaMARCM-F000335_seg001 | Cha-Gal4 | F | 10114.31 | 0.75 |
| Cha-F-000070 | 7546 | ChaMARCM-F000399_seg001 | Cha-Gal4 | F | 10153.51 | 0.76 |
| Cha-F-200046 | 8236 | ChaMARCM-F000233_seg001 | Cha-Gal4 | F | 9904.01 | 0.75 |
| Cha-F-700086 | 8342 | ChaMARCM-F000294_seg001 | Cha-Gal4 | F | 10658.56 | 0.80 |
| Gad1-F-400064 | 9471 | GadMARCM-F000371_seg001 | Gad1-Gal4 | F | 10478.15 | 0.78 |
| Gad1-F-500083 | 9700 | GadMARCM-F000545_seg001 | Gad1-Gal4 | F | 10216.91 | 0.76 |
| Cha-F-400013 | 10082 | ChaMARCM-F000042_seg001 | Cha-Gal4 | F | 10575.30 | 0.80 |
| Gad1-F-600018 | 10592 | GadMARCM-F000108_seg001 | Gad1-Gal4 | F | 10286.69 | 0.79 |
| Gad1-F-900011 | 10622 | GadMARCM-F000125_seg001 | Gad1-Gal4 | F | 6938.39 | 0.53 |
| Gad1-F-200018 | 10709 | GadMARCM-F000042_seg001 | Gad1-Gal4 | F | 10271.59 | 0.77 |
| Gad1-F-400035 | 11364 | GadMARCM-F000321_seg001 | Gad1-Gal4 | F | 9833.70 | 0.73 |
| Gad1-F-200008 | 12336 | GadMARCM-F000024_seg001 | Gad1-Gal4 | F | 10435.09 | 0.77 |
| Gad1-F-100004 | 13169 | GadMARCM-F006_seg1 | Gad1-Gal4 | F | 10640.42 | 0.79 |