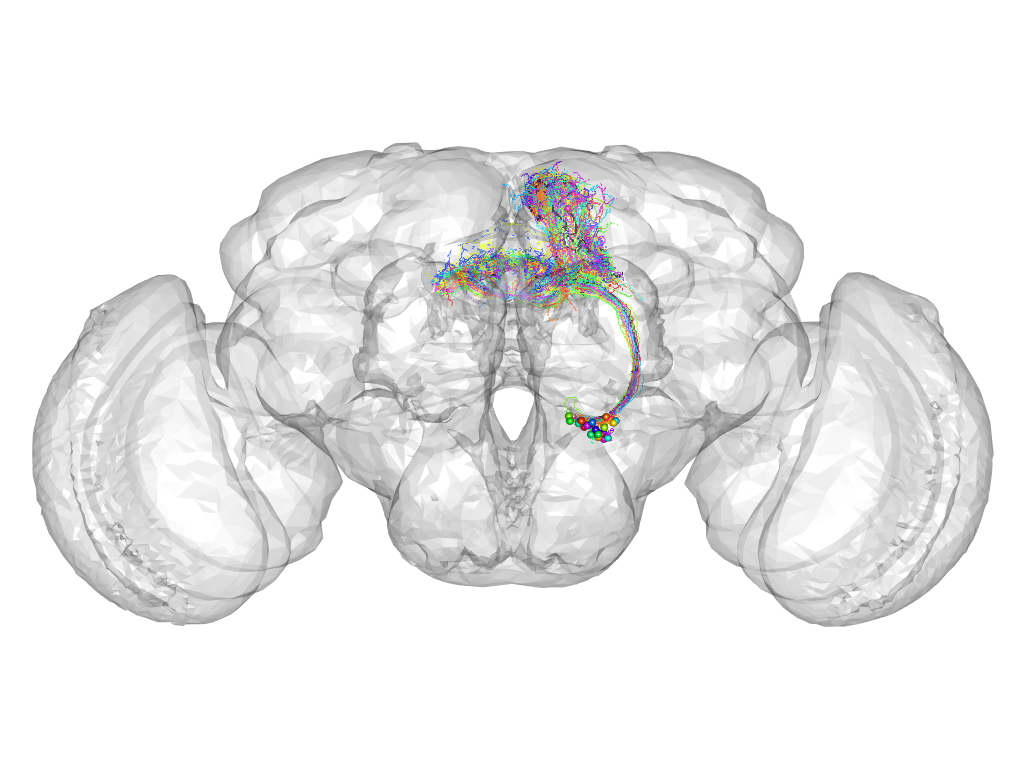
Cluster 142
Part of supercluster XIV
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-400110), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 1028 | VGlut-F-000067 | XIV | 0.67 |
| 204 | Trh-M-100025 | XIV | 0.62 |
| 192 | TH-M-300027 | XIV | 0.58 |
| 859 | TH-F-200089 | XIV | 0.40 |
| 80 | fru-M-400195 | XIV | 0.31 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (Trh-M-400110) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-400110 | 2072 | TPHMARCM-M001575_seg001 | Trh-Gal4 | M | 19020.01 | 1.00 |
| Trh-M-400112 | 2074 | TPHMARCM-M001577_seg001 | Trh-Gal4 | M | 13585.60 | 0.72 |
| Trh-M-400131 | 2148 | TPHMARCM-M001637_seg001 | Trh-Gal4 | M | 13464.48 | 0.69 |
| Trh-M-500162 | 2554 | TPHMARCM-M001529_seg002 | Trh-Gal4 | M | 13875.28 | 0.71 |
| Trh-M-300138 | 2565 | TPHMARCM-M001542_seg001 | Trh-Gal4 | M | 13163.09 | 0.67 |
| Trh-M-300140 | 2567 | TPHMARCM-M001544_seg001 | Trh-Gal4 | M | 12658.80 | 0.66 |
| Trh-M-300134 | 2727 | TPHMARCM-M001491_seg001 | Trh-Gal4 | M | 13808.61 | 0.64 |
| Trh-M-400052 | 2802 | TPHMARCM-1030M_seg1 | Trh-Gal4 | M | 13317.27 | 0.68 |
| Trh-M-400087 | 2859 | TPHMARCM-1088M_seg1 | Trh-Gal4 | M | 11815.54 | 0.67 |
| Trh-M-200055 | 2916 | TPHMARCM-1167M_seg1 | Trh-Gal4 | M | 12543.21 | 0.62 |
| Trh-M-300086 | 2971 | TPHMARCM-911M_seg1 | Trh-Gal4 | M | 12938.56 | 0.70 |
| Trh-M-400049 | 3031 | TPHMARCM-984M_seg1 | Trh-Gal4 | M | 12420.34 | 0.67 |
| Trh-M-300006 | 3463 | TPHMARCM-333M_seg1 | Trh-Gal4 | M | 13144.74 | 0.70 |
| Trh-M-300050 | 3568 | TPHMARCM-499M_seg1 | Trh-Gal4 | M | 13313.86 | 0.69 |
| Trh-M-400023 | 3585 | TPHMARCM-528M_seg1 | Trh-Gal4 | M | 13065.41 | 0.70 |
| Trh-M-300071 | 3687 | TPHMARCM-799M_seg2 | Trh-Gal4 | M | 13673.11 | 0.66 |
| VGlut-F-400851 | 6873 | DvGlutMARCM-F004035_seg001 | VGlut-Gal4 | F | 12864.85 | 0.65 |
| Cha-F-300076 | 8301 | ChaMARCM-F000270_seg002 | Cha-Gal4 | F | 9132.79 | 0.59 |
| Trh-F-200119 | 12403 | TPHMARCM-1198F_seg2 | Trh-Gal4 | F | 12375.92 | 0.62 |
| Trh-F-300146 | 12523 | TPHMARCM-1305F_seg1 | Trh-Gal4 | F | 10894.37 | 0.60 |
| Trh-F-500178 | 12536 | TPHMARCM-1319F_seg2 | Trh-Gal4 | F | 13504.61 | 0.71 |
| Trh-F-200105 | 12697 | TPHMARCM-1146F_seg1 | Trh-Gal4 | F | 13397.50 | 0.68 |
| Trh-F-300137 | 12705 | TPHMARCM-1162F_seg1 | Trh-Gal4 | F | 13789.38 | 0.71 |
| Trh-F-400028 | 13943 | TPHMARCM-235F-repeated0811-06_seg1 | Trh-Gal4 | F | 13618.59 | 0.69 |
| Trh-F-400054 | 14142 | TPHMARCM-554F_seg1 | Trh-Gal4 | F | 13303.64 | 0.71 |
| Trh-F-400055 | 14143 | TPHMARCM-555F_seg1 | Trh-Gal4 | F | 14102.70 | 0.73 |
| Trh-F-300112 | 14282 | TPHMARCM-826F_seg1 | Trh-Gal4 | F | 12612.41 | 0.69 |
| Trh-F-300119 | 14300 | TPHMARCM-839F_seg1 | Trh-Gal4 | F | 12742.03 | 0.67 |