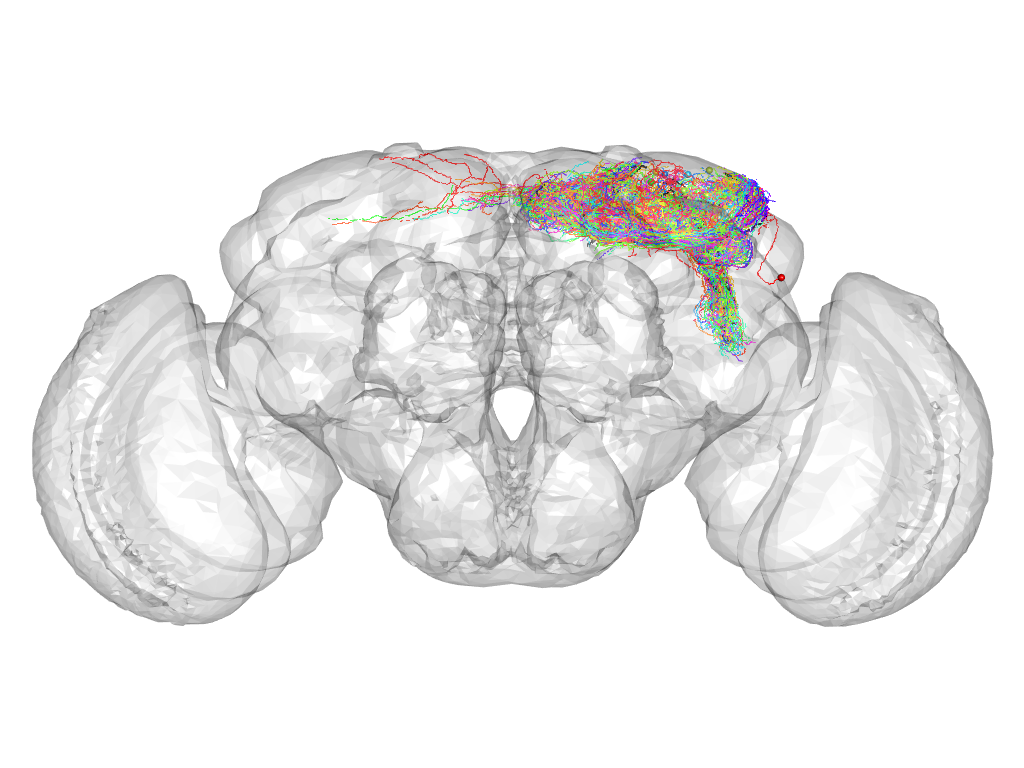
Cluster 100
Part of supercluster XVII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-300168), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 461 | Cha-F-900029 | XVII | 0.57 |
| 28 | fru-M-700179 | XVII | 0.47 |
| 282 | Cha-F-100339 | XVII | 0.45 |
| 65 | fru-M-000139 | XVII | 0.43 |
| 268 | Cha-F-200340 | X | 0.34 |
Cluster composition
This cluster has 55 neurons. The exemplar of this cluster (fru-M-300168) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-300168 | 1305 | FruMARCM-M001515_seg002 | fru-Gal4 | M | 31673.45 | 1.00 |
| fru-M-200274 | 97 | FruMARCM-M002296_seg001 | fru-Gal4 | M | 15827.43 | 0.41 |
| fru-M-000188 | 240 | FruMARCM-M002455_seg001 | fru-Gal4 | M | 21925.03 | 0.67 |
| fru-M-400156 | 507 | FruMARCM-M001611_seg001 | fru-Gal4 | M | 23000.28 | 0.68 |
| fru-M-300231 | 766 | FruMARCM-M001980_seg001 | fru-Gal4 | M | 22667.37 | 0.72 |
| fru-M-000138 | 787 | FruMARCM-M002002_seg002 | fru-Gal4 | M | 22605.79 | 0.71 |
| fru-M-500203 | 801 | FruMARCM-M002011_seg001 | fru-Gal4 | M | 22618.95 | 0.68 |
| fru-M-300244 | 878 | FruMARCM-M002143_seg001 | fru-Gal4 | M | 22294.90 | 0.70 |
| fru-M-700136 | 887 | FruMARCM-M002157_seg001 | fru-Gal4 | M | 22344.35 | 0.70 |
| fru-M-000057 | 1147 | FruMARCM-M001341_seg001 | fru-Gal4 | M | 22186.58 | 0.71 |
| fru-M-200190 | 1214 | FruMARCM-M001399_seg001 | fru-Gal4 | M | 21575.75 | 0.71 |
| fru-M-500072 | 1386 | FruMARCM-M000891_seg001 | fru-Gal4 | M | 20231.80 | 0.67 |
| fru-M-500074 | 1388 | FruMARCM-M000892_seg002 | fru-Gal4 | M | 21627.26 | 0.70 |
| fru-M-000017 | 1560 | FruMARCM-M000630_seg002 | fru-Gal4 | M | 20169.28 | 0.67 |
| fru-F-000121 | 3746 | FruMARCM-F002338_seg001 | fru-Gal4 | F | 23055.15 | 0.71 |
| fru-F-100085 | 3767 | FruMARCM-F002361_seg001 | fru-Gal4 | F | 21869.44 | 0.69 |
| fru-F-400162 | 4498 | FruMARCM-F001601_seg001 | fru-Gal4 | F | 23504.11 | 0.74 |
| fru-F-200143 | 4697 | FruMARCM-F001947_seg001 | fru-Gal4 | F | 21781.83 | 0.69 |
| fru-F-800059 | 4748 | FruMARCM-F002069_seg001 | fru-Gal4 | F | 22339.12 | 0.71 |
| VGlut-F-000009 | 6074 | DvGlutMARCM-F062-X3_seg1 | VGlut-Gal4 | F | 21760.99 | 0.65 |
| fru-F-100012 | 6154 | FruMARCM-F000382_seg003 | fru-Gal4 | F | 13781.01 | 0.59 |
| fru-F-200084 | 6252 | FruMARCM-F001270_seg001 | fru-Gal4 | F | 20968.32 | 0.67 |
| fru-F-000051 | 6290 | FruMARCM-F001386_seg001 | fru-Gal4 | F | 22359.06 | 0.72 |
| fru-F-400158 | 6366 | FruMARCM-F001506_seg002 | fru-Gal4 | F | 20938.78 | 0.69 |
| VGlut-F-800280 | 6581 | DvGlutMARCM-F004342_seg001 | VGlut-Gal4 | F | 20480.30 | 0.67 |
| VGlut-F-700548 | 6644 | DvGlutMARCM-F004391_seg001 | VGlut-Gal4 | F | 20538.65 | 0.69 |
| VGlut-F-300577 | 6895 | DvGlutMARCM-F004051_seg001 | VGlut-Gal4 | F | 22410.47 | 0.69 |
| fru-F-700112 | 7378 | FruMARCM-F001004_seg002 | fru-Gal4 | F | 18317.51 | 0.65 |
| VGlut-F-500730 | 7421 | DvGlutMARCM-F003876_seg001 | VGlut-Gal4 | F | 21535.81 | 0.70 |
| VGlut-F-000594 | 7951 | DvGlutMARCM-F003782_seg002 | VGlut-Gal4 | F | 19783.60 | 0.67 |
| Cha-F-400062 | 8382 | ChaMARCM-F000322_seg001 | Cha-Gal4 | F | 20553.35 | 0.67 |
| VGlut-F-700405 | 8619 | DvGlutMARCM-F003673_seg001 | VGlut-Gal4 | F | 22256.13 | 0.72 |
| VGlut-F-000534 | 8728 | DvGlutMARCM-F003559_seg002 | VGlut-Gal4 | F | 19201.05 | 0.65 |
| Gad1-F-100039 | 9577 | GadMARCM-F000448_seg001 | Gad1-Gal4 | F | 9567.74 | 0.48 |
| fru-F-100008 | 9790 | FruMARCM-F000309_seg001 | fru-Gal4 | F | 22203.51 | 0.72 |
| fru-F-200035 | 9851 | FruMARCM-F000439_seg001 | fru-Gal4 | F | 21690.12 | 0.70 |
| VGlut-F-200437 | 11056 | DvGlutMARCM-F002935_seg001 | VGlut-Gal4 | F | 21754.89 | 0.70 |
| VGlut-F-500426 | 12050 | DvGlutMARCM-F002564_seg001 | VGlut-Gal4 | F | 21863.35 | 0.70 |
| VGlut-F-700031 | 12763 | DvGlutMARCM-F1817_seg1 | VGlut-Gal4 | F | 21798.47 | 0.72 |
| VGlut-F-400456 | 12862 | DvGlutMARCM-F1903_seg2 | VGlut-Gal4 | F | 19692.56 | 0.64 |
| VGlut-F-300399 | 13161 | DvGlutMARCM-F2198_seg1 | VGlut-Gal4 | F | 21849.43 | 0.69 |
| VGlut-F-000287 | 14397 | DvGlutMARCM-F1077_seg1 | VGlut-Gal4 | F | 18895.48 | 0.63 |
| VGlut-F-200235 | 14836 | DvGlutMARCM-F1180_seg1 | VGlut-Gal4 | F | 17694.09 | 0.63 |
| VGlut-F-900022 | 14975 | DvGlutMARCM-F1314_seg1 | VGlut-Gal4 | F | 21372.89 | 0.71 |
| VGlut-F-300240 | 15011 | DvGlutMARCM-F1351_seg1 | VGlut-Gal4 | F | 19625.54 | 0.65 |
| VGlut-F-800033 | 15130 | DvGlutMARCM-F1463_seg1 | VGlut-Gal4 | F | 18374.77 | 0.65 |
| VGlut-F-300291 | 15238 | DvGlutMARCM-F1560_seg1 | VGlut-Gal4 | F | 22352.27 | 0.71 |
| VGlut-F-200003 | 15591 | DvGlutMARCM-F006_seg2 | VGlut-Gal4 | F | 21869.59 | 0.70 |
| VGlut-F-400005 | 15641 | DvGlutMARCM-F055_seg1 | VGlut-Gal4 | F | 18811.49 | 0.64 |
| VGlut-F-300008 | 15680 | DvGlutMARCM-F095_seg1 | VGlut-Gal4 | F | 21890.17 | 0.69 |
| VGlut-F-100008 | 15760 | DvGlutMARCM-F167_seg1 | VGlut-Gal4 | F | 21792.39 | 0.71 |
| VGlut-F-600004 | 15808 | DvGlutMARCM-F216_seg1 | VGlut-Gal4 | F | 21882.67 | 0.71 |
| VGlut-F-400034 | 15810 | DvGlutMARCM-F218_seg1 | VGlut-Gal4 | F | 19586.56 | 0.64 |
| VGlut-F-300070 | 15968 | DvGlutMARCM-F360_seg1 | VGlut-Gal4 | F | 20907.70 | 0.66 |
| VGlut-F-300084 | 16160 | DvGlutMARCM-F542_seg1 | VGlut-Gal4 | F | 20989.32 | 0.70 |