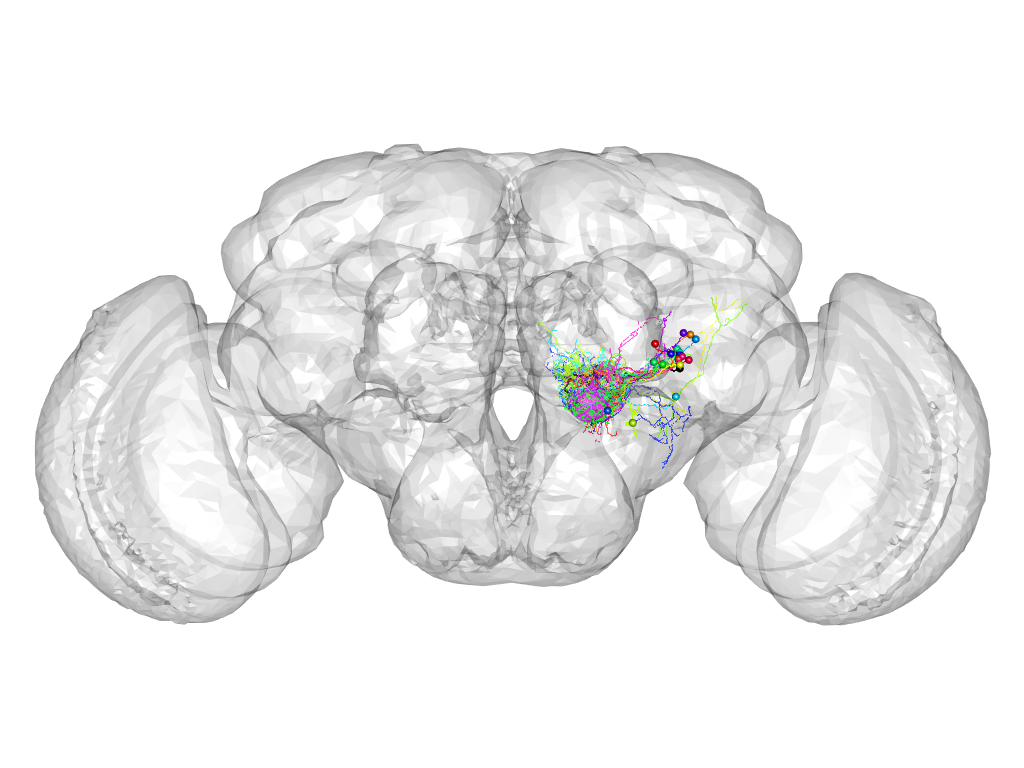
Cluster 953
Part of supercluster XII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-000356), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 806 | Trh-F-100085 | XII | 0.75 |
| 824 | VGlut-F-200354 | XII | 0.67 |
| 545 | VGlut-F-700278 | XII | 0.66 |
| 513 | Cha-F-300036 | XII | 0.66 |
| 748 | 5-HT1B-F-500012 | XII | 0.65 |
| 835 | VGlut-F-400504 | XII | 0.64 |
| 653 | Trh-F-000043 | XIX | 0.63 |
| 926 | VGlut-F-500134 | XII | 0.61 |
| 872 | TH-F-100101 | IX | 0.60 |
| 932 | VGlut-F-100102 | XII | 0.60 |
| 144 | Trh-M-400113 | XII | 0.58 |
| 662 | Tdc2-F-400026 | XIII | 0.58 |
| 201 | Trh-M-100007 | XII | 0.57 |
| 666 | TH-F-200011 | IX | 0.55 |
| 400 | VGlut-F-500072 | XII | 0.54 |
| 258 | Cha-F-100277 | XIII | 0.54 |
Cluster composition
This cluster has 24 neurons. The exemplar of this cluster (VGlut-F-000356) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-000356 | 15065 | DvGlutMARCM-F1402_seg1 | VGlut-Gal4 | F | 8621.65 | 1.00 |
| fru-M-500189 | 727 | FruMARCM-M001937_seg001 | fru-Gal4 | M | 6459.10 | 0.73 |
| fru-M-600052 | 1180 | FruMARCM-M001363_seg001 | fru-Gal4 | M | 6203.64 | 0.74 |
| fru-M-800023 | 1593 | FruMARCM-M000663_seg001 | fru-Gal4 | M | 6073.28 | 0.72 |
| fru-M-200037 | 2447 | FruMARCM-M000209_seg003 | fru-Gal4 | M | 6316.11 | 0.73 |
| Trh-M-200074 | 2648 | TPHMARCM-M001417_seg001 | Trh-Gal4 | M | 6104.86 | 0.50 |
| TH-M-400007 | 3139 | THMARCM-181M_seg2 | TH-Gal4 | M | 5975.72 | 0.60 |
| Cha-F-100123 | 4855 | ChaMARCM-F000977_seg002 | Cha-Gal4 | F | 5769.90 | 0.49 |
| Cha-F-000210 | 5015 | ChaMARCM-F001105_seg001 | Cha-Gal4 | F | 6559.69 | 0.74 |
| Cha-F-600146 | 5643 | ChaMARCM-F000920_seg001 | Cha-Gal4 | F | 5522.55 | 0.59 |
| VGlut-F-400508 | 6096 | DvGlutMARCM-F2126_seg2 | VGlut-Gal4 | F | 5815.10 | 0.69 |
| Gad1-F-300076 | 6147 | GadMARCM-F000326_seg001 | Gad1-Gal4 | F | 5715.61 | 0.68 |
| fru-F-000065 | 6378 | FruMARCM-F001514_seg002 | fru-Gal4 | F | 6413.27 | 0.73 |
| VGlut-F-300625 | 6494 | DvGlutMARCM-F004272_seg001 | VGlut-Gal4 | F | 5793.55 | 0.61 |
| VGlut-F-000622 | 6889 | DvGlutMARCM-F004045_seg001 | VGlut-Gal4 | F | 6179.15 | 0.71 |
| Cha-F-600088 | 7742 | ChaMARCM-F000547_seg003 | Cha-Gal4 | F | 3738.67 | 0.55 |
| Gad1-F-100049 | 9608 | GadMARCM-F000472_seg002 | Gad1-Gal4 | F | 5277.87 | 0.45 |
| fru-F-000005 | 11539 | FruMARCM-F000196_seg002 | fru-Gal4 | F | 5751.37 | 0.64 |
| VGlut-F-000284 | 14394 | DvGlutMARCM-F1074_seg2 | VGlut-Gal4 | F | 6452.79 | 0.74 |
| VGlut-F-000237 | 14712 | DvGlutMARCM-F971_seg1 | VGlut-Gal4 | F | 6008.42 | 0.65 |
| VGlut-F-000248 | 14724 | DvGlutMARCM-F984_seg1 | VGlut-Gal4 | F | 5682.39 | 0.61 |
| VGlut-F-100075 | 14731 | DvGlutMARCM-F991_seg1 | VGlut-Gal4 | F | 5308.41 | 0.58 |
| VGlut-F-100172 | 15333 | DvGlutMARCM-F1678_seg1 | VGlut-Gal4 | F | 6358.72 | 0.73 |
| VGlut-F-100192 | 15411 | DvGlutMARCM-F1769_seg1 | VGlut-Gal4 | F | 6468.71 | 0.77 |