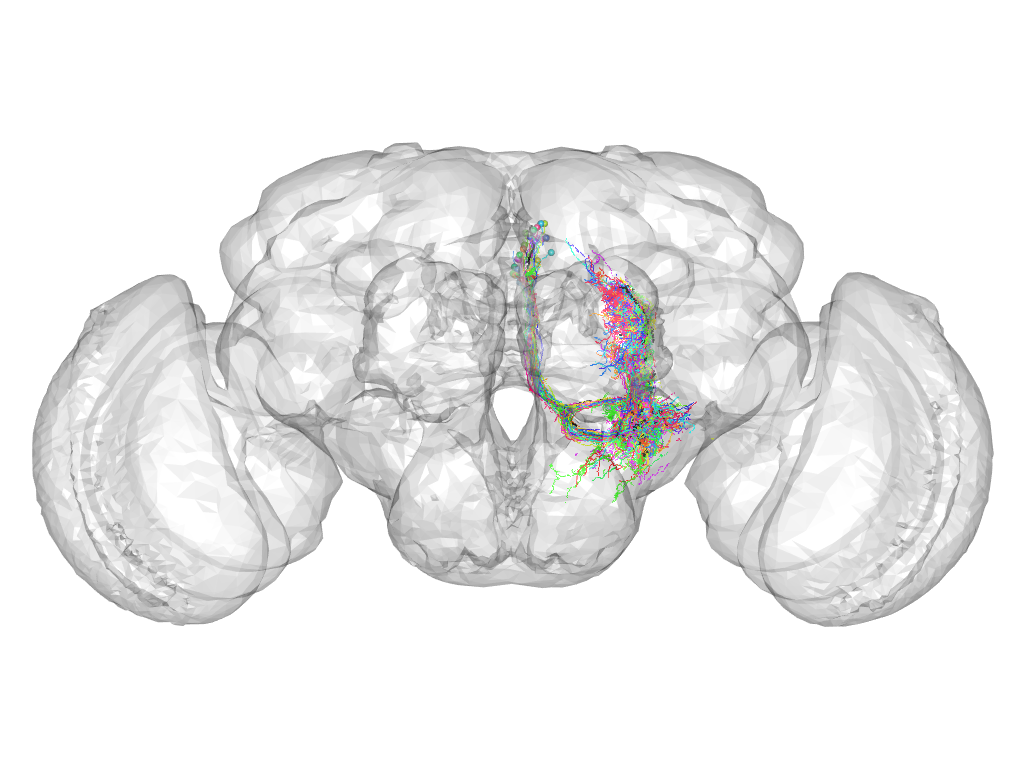
Cluster 921
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-300181), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 187 | TH-M-400003 | VIII | 0.67 |
| 855 | TH-F-200057 | VIII | 0.66 |
| 406 | fru-F-000053 | VII | 0.60 |
| 667 | TH-F-300071 | XIV | 0.59 |
| 686 | VGlut-F-900065 | VII | 0.52 |
| 988 | VGlut-F-200005 | X | 0.52 |
| 852 | TH-F-100030 | VII | 0.50 |
| 191 | TH-M-200018 | XIV | 0.50 |
Cluster composition
This cluster has 33 neurons. The exemplar of this cluster (VGlut-F-300181) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-300181 | 14598 | DvGlutMARCM-F860_seg1 | VGlut-Gal4 | F | 36081.08 | 1.00 |
| VGlut-F-400910 | 5810 | DvGlutMARCM-F004506_seg002 | VGlut-Gal4 | F | 23580.78 | 0.61 |
| VGlut-F-700543 | 6626 | DvGlutMARCM-F004378_seg001 | VGlut-Gal4 | F | 24177.46 | 0.68 |
| VGlut-F-500778 | 7070 | DvGlutMARCM-F004199_seg002 | VGlut-Gal4 | F | 26639.89 | 0.73 |
| VGlut-F-200471 | 9217 | DvGlutMARCM-F003283_seg001 | VGlut-Gal4 | F | 24110.61 | 0.61 |
| VGlut-F-400665 | 9348 | DvGlutMARCM-F003179_seg001 | VGlut-Gal4 | F | 26134.12 | 0.71 |
| VGlut-F-600164 | 10213 | DvGlutMARCM-F002300_seg002 | VGlut-Gal4 | F | 25678.12 | 0.72 |
| VGlut-F-000281 | 10759 | DVGlutMARCM-F-0625-07-F1073_seg2 | VGlut-Gal4 | F | 22040.73 | 0.64 |
| VGlut-F-600290 | 10775 | DvGlutMARCM-F002537_seg001 | VGlut-Gal4 | F | 25077.65 | 0.72 |
| VGlut-F-700190 | 10914 | DvGlutMARCM-F002814_seg001 | VGlut-Gal4 | F | 23946.01 | 0.70 |
| VGlut-F-600499 | 11650 | DvGlutMARCM-F002224_seg001 | VGlut-Gal4 | F | 25624.81 | 0.72 |
| VGlut-F-600504 | 11659 | DvGlutMARCM-F002233_seg001 | VGlut-Gal4 | F | 22994.20 | 0.60 |
| VGlut-F-400640 | 11666 | DvGlutMARCM-F002245_seg001 | VGlut-Gal4 | F | 24206.94 | 0.68 |
| VGlut-F-600181 | 11744 | DvGlutMARCM-F002316_seg001 | VGlut-Gal4 | F | 25953.63 | 0.70 |
| VGlut-F-600182 | 11745 | DvGlutMARCM-F002317_seg001 | VGlut-Gal4 | F | 23786.77 | 0.68 |
| VGlut-F-400545 | 11948 | DvGlutMARCM-F002483_seg001 | VGlut-Gal4 | F | 26347.46 | 0.74 |
| VGlut-F-500421 | 12012 | DvGlutMARCM-F002534_seg001 | VGlut-Gal4 | F | 24009.64 | 0.61 |
| VGlut-F-600115 | 12948 | DvGlutMARCM-F1980_seg3 | VGlut-Gal4 | F | 25258.29 | 0.69 |
| VGlut-F-400507 | 13150 | DvGlutMARCM-F2183_seg1 | VGlut-Gal4 | F | 24090.51 | 0.68 |
| VGlut-F-300196 | 14369 | DvGlutMARCM-F1050_seg1 | VGlut-Gal4 | F | 24162.80 | 0.72 |
| VGlut-F-300200 | 14411 | DvGlutMARCM-F1091_seg1 | VGlut-Gal4 | F | 24847.05 | 0.63 |
| VGlut-F-300142 | 14425 | DvGlutMARCM-F719_seg1 | VGlut-Gal4 | F | 24160.23 | 0.70 |
| VGlut-F-400194 | 14465 | DvGlutMARCM-F748_seg1 | VGlut-Gal4 | F | 23742.26 | 0.67 |
| VGlut-F-400206 | 14486 | DvGlutMARCM-F760-x2_seg3 | VGlut-Gal4 | F | 25235.28 | 0.63 |
| VGlut-F-400227 | 14536 | DvGlutMARCM-F806-x2_seg1 | VGlut-Gal4 | F | 12364.00 | 0.56 |
| VGlut-F-300223 | 14873 | DvGlutMARCM-F1217_seg1 | VGlut-Gal4 | F | 24092.83 | 0.69 |
| VGlut-F-500147 | 14942 | DvGlutMARCM-F1280_seg2 | VGlut-Gal4 | F | 25616.68 | 0.70 |
| VGlut-F-000364 | 15140 | DvGlutMARCM-F1473_seg1 | VGlut-Gal4 | F | 23670.16 | 0.62 |
| VGlut-F-300288 | 15235 | DvGlutMARCM-F1558_seg1 | VGlut-Gal4 | F | 26209.29 | 0.72 |
| VGlut-F-500244 | 15393 | DvGlutMARCM-F1751_seg2 | VGlut-Gal4 | F | 24436.01 | 0.69 |
| VGlut-F-400129 | 15515 | DvGlutMARCM-F663-X2_seg1 | VGlut-Gal4 | F | 24515.19 | 0.68 |
| VGlut-F-200084 | 15868 | DvGlutMARCM-F271_seg1 | VGlut-Gal4 | F | 23806.84 | 0.63 |
| VGlut-F-300035 | 15911 | DvGlutMARCM-F309_seg1 | VGlut-Gal4 | F | 25004.28 | 0.61 |