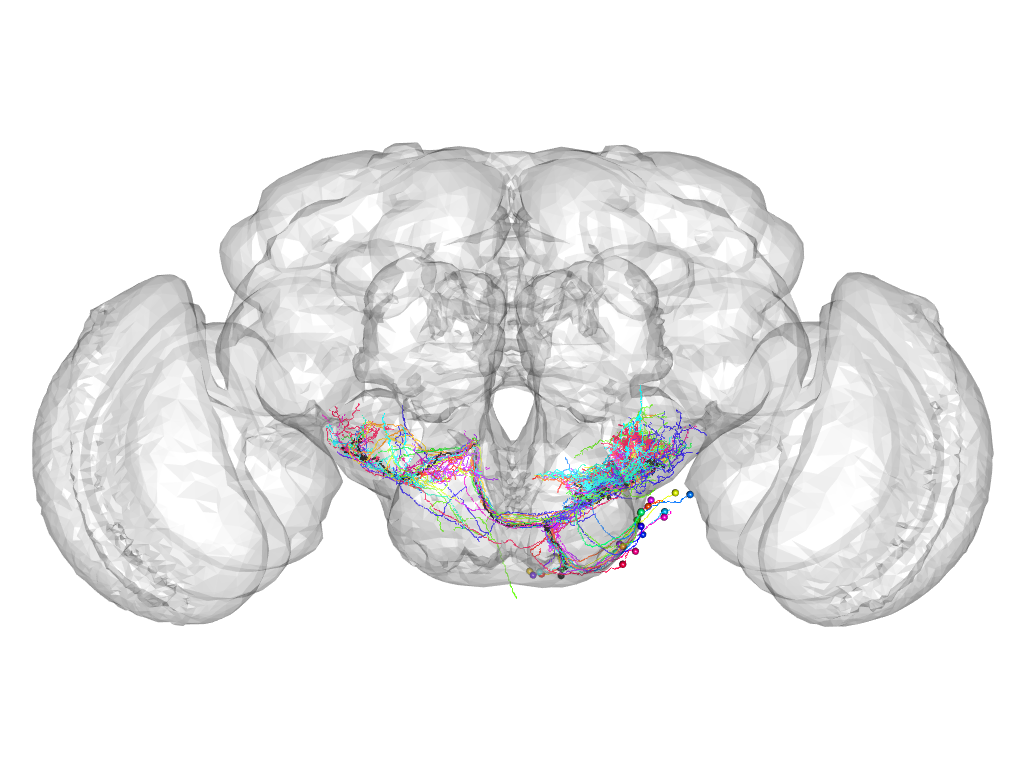
Cluster 918
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-200157), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 546 | VGlut-F-000492 | VIII | 0.62 |
| 652 | Tdc2-F-200010 | VIII | 0.60 |
| 9 | fru-M-700156 | VIII | 0.45 |
| 302 | fru-F-000090 | VII | 0.41 |
| 690 | VGlut-F-400061 | VIII | 0.36 |
Cluster composition
This cluster has 23 neurons. The exemplar of this cluster (VGlut-F-200157) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-200157 | 14566 | DvGlutMARCM-F831_seg1 | VGlut-Gal4 | F | 31855.68 | 1.00 |
| fru-M-200336 | 12 | FruMARCM-M002591_seg001 | fru-Gal4 | M | 19020.52 | 0.57 |
| fru-M-100205 | 19 | FruMARCM-M002206_seg001 | fru-Gal4 | M | 21431.67 | 0.63 |
| fru-M-400226 | 390 | FruMARCM-M002623_seg001 | fru-Gal4 | M | 19957.86 | 0.59 |
| fru-M-100155 | 479 | FruMARCM-M001582_seg001 | fru-Gal4 | M | 21778.39 | 0.67 |
| fru-M-300227 | 739 | FruMARCM-M001951_seg001 | fru-Gal4 | M | 20484.26 | 0.62 |
| fru-M-200240 | 836 | FruMARCM-M002045_seg001 | fru-Gal4 | M | 19971.60 | 0.63 |
| fru-M-400119 | 1009 | FruMARCM-M001187_seg002 | fru-Gal4 | M | 13806.29 | 0.30 |
| fru-M-300104 | 1456 | FruMARCM-M001021_seg002 | fru-Gal4 | M | 16380.42 | 0.57 |
| fru-M-300003 | 2344 | FruMARCM-M000033_seg002 | fru-Gal4 | M | 21533.74 | 0.68 |
| Cha-F-300007 | 6141 | ChaMARCM-F000013_seg001 | Cha-Gal4 | F | 11967.17 | 0.45 |
| VGlut-F-500802 | 6524 | DvGlutMARCM-F004294_seg003 | VGlut-Gal4 | F | 21955.46 | 0.61 |
| VGlut-F-600720 | 6794 | DvGlutMARCM-F003987_seg001 | VGlut-Gal4 | F | 23412.22 | 0.59 |
| VGlut-F-000648 | 7058 | DvGlutMARCM-F004187_seg001 | VGlut-Gal4 | F | 21126.04 | 0.65 |
| VGlut-F-300512 | 8584 | DvGlutMARCM-F003651_seg001 | VGlut-Gal4 | F | 11675.62 | 0.41 |
| VGlut-F-500332 | 13038 | DvGlutMARCM-F2062_seg1 | VGlut-Gal4 | F | 21903.35 | 0.69 |
| VGlut-F-500124 | 14609 | DvGlutMARCM-F871_seg1 | VGlut-Gal4 | F | 13590.46 | 0.29 |
| VGlut-F-000224 | 14694 | DvGlutMARCM-F952_seg1 | VGlut-Gal4 | F | 21664.40 | 0.68 |
| VGlut-F-400283 | 14696 | DvGlutMARCM-F953_seg2 | VGlut-Gal4 | F | 18931.52 | 0.55 |
| VGlut-F-100081 | 14737 | DvGlutMARCM-F995_seg3 | VGlut-Gal4 | F | 17616.07 | 0.55 |
| VGlut-F-300098 | 15443 | DvGlutMARCM-F601_seg2 | VGlut-Gal4 | F | 21789.92 | 0.65 |
| VGlut-F-500064 | 16026 | DvGlutMARCM-F417_seg1 | VGlut-Gal4 | F | 18526.87 | 0.59 |
| VGlut-F-000151 | 16173 | DvGlutMARCM-F555_seg1 | VGlut-Gal4 | F | 17928.25 | 0.45 |