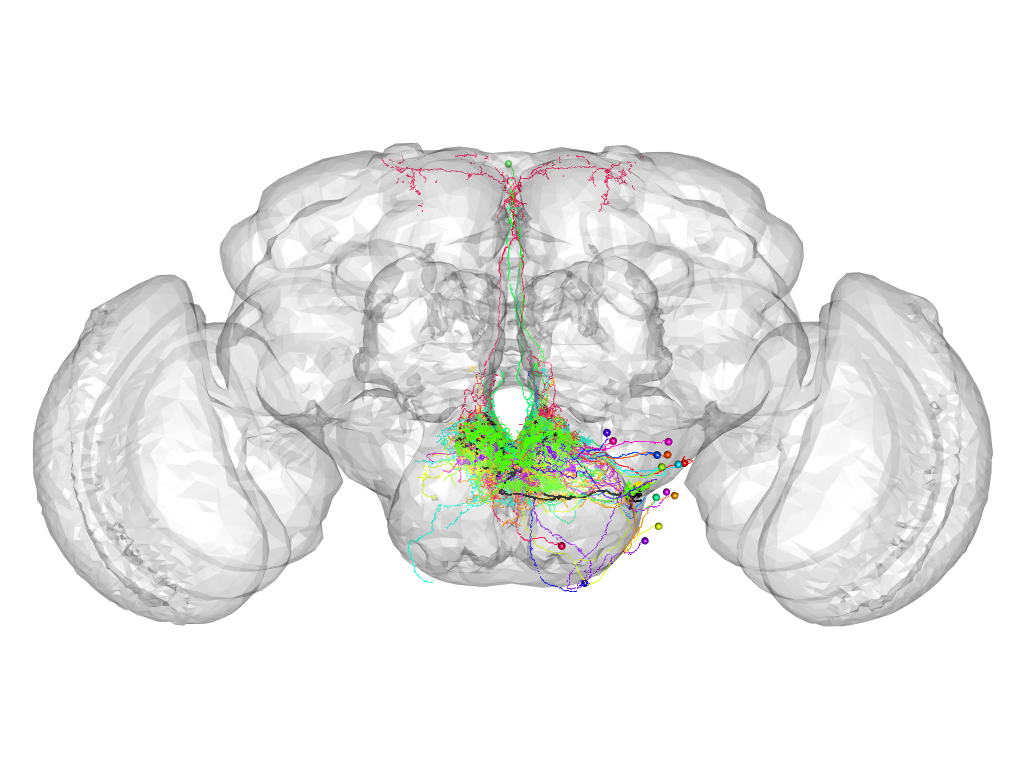
Cluster 868
Part of supercluster IX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-F-000049), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 687 | VGlut-F-500173 | IX | 0.65 |
| 206 | Trh-M-000018 | XIX | 0.47 |
| 152 | Trh-M-000122 | IX | 0.43 |
| 283 | Cha-F-100343 | IX | 0.39 |
| 698 | VGlut-F-600430 | IX | 0.32 |
Cluster composition
This cluster has 22 neurons. The exemplar of this cluster (TH-F-000049) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-F-000049 | 13659 | THMARCM-672F_seg1 | TH-Gal4 | F | 160246.46 | 1.00 |
| fru-M-000162 | 31 | FruMARCM-M002230_seg001 | fru-Gal4 | M | 101418.80 | 0.67 |
| fru-M-200282 | 119 | FruMARCM-M002315_seg002 | fru-Gal4 | M | 96243.21 | 0.62 |
| fru-M-200114 | 1512 | FruMARCM-M001060_seg002 | fru-Gal4 | M | 96412.05 | 0.58 |
| TH-M-000037 | 2015 | THMARCM-609M_seg | TH-Gal4 | M | 125997.43 | 0.78 |
| Trh-M-000054 | 2061 | TPHMARCM-M001566_seg001 | Trh-Gal4 | M | 67722.63 | 0.48 |
| fru-M-200036 | 2446 | FruMARCM-M000209_seg002 | fru-Gal4 | M | 92299.73 | 0.62 |
| TH-M-000014 | 3303 | THMARCM-524M_seg1 | TH-Gal4 | M | 127453.86 | 0.77 |
| VGlut-F-700605 | 5944 | DvGlutMARCM-F004599_seg001 | VGlut-Gal4 | F | 96347.02 | 0.54 |
| Cha-F-500111 | 7706 | ChaMARCM-F000523_seg002 | Cha-Gal4 | F | 107886.84 | 0.61 |
| VGlut-F-400771 | 8706 | DvGlutMARCM-F003741_seg001 | VGlut-Gal4 | F | 107392.32 | 0.66 |
| VGlut-F-100313 | 8844 | DvGlutMARCM-F003638_seg001 | VGlut-Gal4 | F | 112787.04 | 0.62 |
| Cha-F-000016 | 10163 | ChaMARCM-F000094_seg002 | Cha-Gal4 | F | 101470.41 | 0.68 |
| Gad1-F-200096 | 11342 | GadMARCM-F000307_seg002 | Gad1-Gal4 | F | 116507.60 | 0.75 |
| VGlut-F-200359 | 12976 | DvGlutMARCM-F2010_seg2 | VGlut-Gal4 | F | 37482.91 | 0.43 |
| Trh-F-800004 | 13274 | TPHMARCM-759F_seg2 | Trh-Gal4 | F | 97929.98 | 0.68 |
| Trh-F-200037 | 13974 | TPHMARCM-264F_seg1 | Trh-Gal4 | F | 40992.06 | 0.47 |
| VGlut-F-000272 | 14363 | DvGlutMARCM-F1044_seg1 | VGlut-Gal4 | F | 86954.22 | 0.63 |
| VGlut-F-100103 | 14766 | DvGlutMARCM-F1111_seg1 | VGlut-Gal4 | F | 49571.31 | 0.41 |
| VGlut-F-300303 | 15276 | DvGlutMARCM-F1603_seg1 | VGlut-Gal4 | F | 84194.71 | 0.59 |
| VGlut-F-400016 | 15704 | DvGlutMARCM-F120_seg1 | VGlut-Gal4 | F | 86509.82 | 0.66 |
| VGlut-F-000164 | 16191 | DvGlutMARCM-F573_seg1 | VGlut-Gal4 | F | 117757.48 | 0.55 |