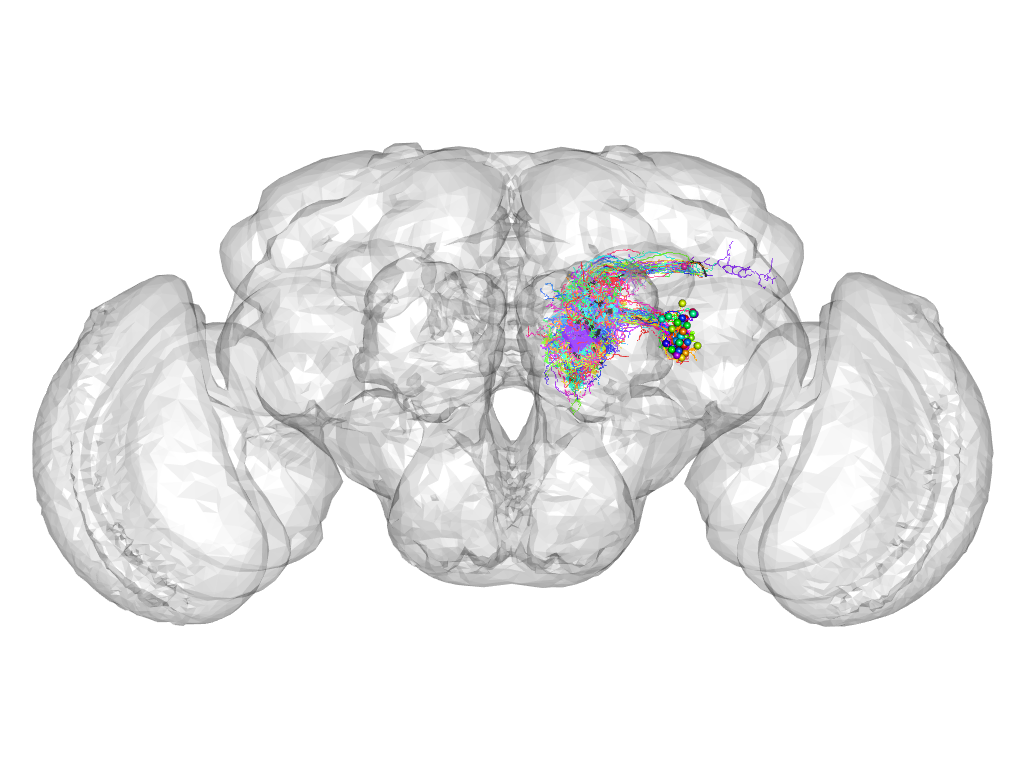
Cluster 821
Part of supercluster XI
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-600111), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 1051 | VGlut-F-200115 | XI | 0.75 |
| 662 | Tdc2-F-400026 | XIII | 0.70 |
| 421 | VGlut-F-700518 | XI | 0.65 |
| 806 | Trh-F-100085 | XII | 0.64 |
| 144 | Trh-M-400113 | XII | 0.64 |
| 201 | Trh-M-100007 | XII | 0.62 |
| 400 | VGlut-F-500072 | XII | 0.57 |
| 914 | VGlut-F-600026 | XI | 0.56 |
| 545 | VGlut-F-700278 | XII | 0.51 |
| 172 | Trh-M-100092 | XII | 0.51 |
Cluster composition
This cluster has 48 neurons. The exemplar of this cluster (VGlut-F-600111) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-600111 | 12934 | DvGlutMARCM-F1971_seg2 | VGlut-Gal4 | F | 28723.64 | 1.00 |
| TH-M-700011 | 3114 | THMARCM-127M_seg2 | TH-Gal4 | M | 13603.28 | 0.47 |
| VGlut-F-800291 | 5733 | DvGlutMARCM-F004440_seg001 | VGlut-Gal4 | F | 18994.75 | 0.67 |
| VGlut-F-900134 | 5800 | DvGlutMARCM-F004498_seg001 | VGlut-Gal4 | F | 19838.24 | 0.72 |
| VGlut-F-700576 | 5805 | DvGlutMARCM-F004502_seg001 | VGlut-Gal4 | F | 19172.41 | 0.70 |
| VGlut-F-100366 | 6415 | DvGlutMARCM-F004207_seg001 | VGlut-Gal4 | F | 20382.76 | 0.70 |
| VGlut-F-500742 | 6740 | DvGlutMARCM-F003945_seg001 | VGlut-Gal4 | F | 18250.76 | 0.65 |
| VGlut-F-100349 | 7429 | DvGlutMARCM-F003883_seg001 | VGlut-Gal4 | F | 19163.94 | 0.69 |
| VGlut-F-900108 | 7962 | DvGlutMARCM-F003792_seg001 | VGlut-Gal4 | F | 13880.67 | 0.59 |
| VGlut-F-700414 | 7963 | DvGlutMARCM-F003793_seg001 | VGlut-Gal4 | F | 14859.77 | 0.60 |
| VGlut-F-900109 | 7983 | DvGlutMARCM-F003808_seg001 | VGlut-Gal4 | F | 18278.96 | 0.65 |
| VGlut-F-500719 | 8594 | DvGlutMARCM-F003659_seg001 | VGlut-Gal4 | F | 18896.19 | 0.67 |
| VGlut-F-900097 | 8635 | DvGlutMARCM-F003688_seg001 | VGlut-Gal4 | F | 18813.26 | 0.68 |
| VGlut-F-900100 | 8644 | DvGlutMARCM-F003694_seg001 | VGlut-Gal4 | F | 19613.66 | 0.68 |
| VGlut-F-500703 | 8773 | DvGlutMARCM-F003592_seg001 | VGlut-Gal4 | F | 19168.47 | 0.66 |
| VGlut-F-600655 | 8971 | DvGlutMARCM-F003531_seg001 | VGlut-Gal4 | F | 17086.31 | 0.67 |
| VGlut-F-700333 | 9067 | DvGlutMARCM-F003385_seg001 | VGlut-Gal4 | F | 19778.00 | 0.70 |
| VGlut-F-700344 | 9109 | DvGlutMARCM-F003415_seg001 | VGlut-Gal4 | F | 20005.00 | 0.70 |
| VGlut-F-600581 | 9259 | DvGlutMARCM-F003318_seg001 | VGlut-Gal4 | F | 14950.34 | 0.60 |
| VGlut-F-800148 | 9266 | DvGlutMARCM-F003324_seg001 | VGlut-Gal4 | F | 20310.97 | 0.72 |
| VGlut-F-600508 | 10220 | DvGlutMARCM-F003040_seg001 | VGlut-Gal4 | F | 17690.33 | 0.69 |
| Gad1-F-600027 | 10523 | GadMARCM-F000164_seg001 | Gad1-Gal4 | F | 15607.20 | 0.56 |
| Gad1-F-300036 | 10564 | GadMARCM-F000087_seg001 | Gad1-Gal4 | F | 17382.16 | 0.65 |
| VGlut-F-300190 | 10749 | DvGlutMARCM-F1022_seg1 | VGlut-Gal4 | F | 21012.17 | 0.72 |
| VGlut-F-800012 | 10764 | DvGlutMARCM-F-0321-01-F193_seg1 | VGlut-Gal4 | F | 21483.99 | 0.72 |
| VGlut-F-700203 | 10784 | DvGlutMARCM-F002832_seg001 | VGlut-Gal4 | F | 18808.12 | 0.68 |
| VGlut-F-700164 | 10891 | DvGlutMARCM-F002797_seg001 | VGlut-Gal4 | F | 19973.82 | 0.70 |
| VGlut-F-900061 | 11071 | DvGlutMARCM-F002950_seg001 | VGlut-Gal4 | F | 20245.85 | 0.71 |
| VGlut-F-700232 | 11114 | DvGlutMARCM-F002980_seg001 | VGlut-Gal4 | F | 19979.81 | 0.69 |
| VGlut-F-600477 | 11122 | DvGlutMARCM-F002987_seg001 | VGlut-Gal4 | F | 20057.08 | 0.70 |
| VGlut-F-800116 | 11155 | DvGlutMARCM-F003008_seg001 | VGlut-Gal4 | F | 19384.59 | 0.68 |
| VGlut-F-700062 | 11873 | DvGlutMARCM-F002418_seg001 | VGlut-Gal4 | F | 18619.66 | 0.68 |
| VGlut-F-700063 | 11876 | DvGlutMARCM-F002420_seg001 | VGlut-Gal4 | F | 18412.46 | 0.67 |
| VGlut-F-700072 | 12078 | DvGlutMARCM-F002587_seg001 | VGlut-Gal4 | F | 18574.42 | 0.68 |
| VGlut-F-500289 | 12818 | DvGlutMARCM-F1866_seg1 | VGlut-Gal4 | F | 13819.54 | 0.57 |
| VGlut-F-600100 | 12911 | DvGlutMARCM-F1952_seg1 | VGlut-Gal4 | F | 19620.89 | 0.70 |
| VGlut-F-600107 | 12926 | DvGlutMARCM-F1964_seg1 | VGlut-Gal4 | F | 13151.86 | 0.48 |
| VGlut-F-300383 | 13018 | DvGlutMARCM-F2044_seg1 | VGlut-Gal4 | F | 17939.44 | 0.65 |
| VGlut-F-600149 | 13147 | DvGlutMARCM-F2179_seg1 | VGlut-Gal4 | F | 19536.69 | 0.70 |
| VGlut-F-400292 | 14339 | DvGlutMARCM-F1021_seg1 | VGlut-Gal4 | F | 18335.51 | 0.68 |
| VGlut-F-600027 | 14479 | DvGlutMARCM-F758_seg1 | VGlut-Gal4 | F | 19181.74 | 0.68 |
| VGlut-F-400245 | 14581 | DvGlutMARCM-F846_seg1 | VGlut-Gal4 | F | 18572.55 | 0.68 |
| VGlut-F-000206 | 14599 | DvGlutMARCM-F861_seg1 | VGlut-Gal4 | F | 18206.85 | 0.60 |
| VGlut-F-500157 | 15043 | DvGlutMARCM-F1381_seg1 | VGlut-Gal4 | F | 20409.31 | 0.69 |
| VGlut-F-800032 | 15125 | DvGlutMARCM-F1458_seg1 | VGlut-Gal4 | F | 16994.38 | 0.65 |
| VGlut-F-600073 | 15152 | DvGlutMARCM-F1485_seg1 | VGlut-Gal4 | F | 19000.67 | 0.67 |
| VGlut-F-600007 | 15848 | DvGlutMARCM-F255_seg1 | VGlut-Gal4 | F | 19338.03 | 0.69 |
| VGlut-F-600009 | 15850 | DvGlutMARCM-F256-X2_seg2 | VGlut-Gal4 | F | 16887.73 | 0.63 |