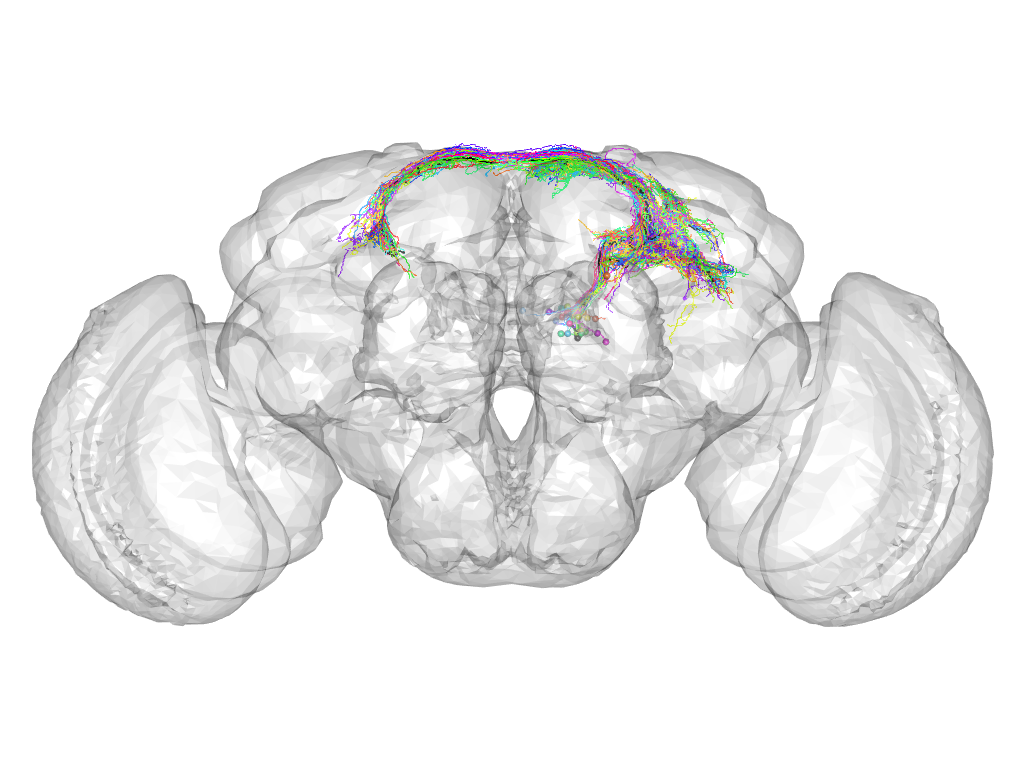
Cluster 81
Part of supercluster XVIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-100203), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 22 | fru-M-300288 | XVIII | 0.76 |
| 117 | fru-M-500059 | XVIII | 0.69 |
| 225 | fru-F-300124 | X | 0.69 |
| 165 | Trh-M-500167 | XVIII | 0.58 |
| 222 | fru-F-000131 | XIX | 0.54 |
| 124 | fru-M-500012 | X | 0.54 |
| 411 | fru-F-700151 | X | 0.54 |
| 90 | fru-M-200156 | X | 0.52 |
| 92 | fru-M-100103 | X | 0.52 |
| 30 | fru-M-000203 | XVIII | 0.51 |
| 91 | fru-M-200161 | IX | 0.51 |
Cluster composition
This cluster has 34 neurons. The exemplar of this cluster (fru-M-100203) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-100203 | 914 | FruMARCM-M002175_seg001 | fru-Gal4 | M | 26502.74 | 1.00 |
| fru-M-000167 | 43 | FruMARCM-M002238_seg001 | fru-Gal4 | M | 19698.52 | 0.74 |
| fru-M-100206 | 57 | FruMARCM-M002253_seg001 | fru-Gal4 | M | 19130.25 | 0.72 |
| fru-M-800092 | 82 | FruMARCM-M002281_seg001 | fru-Gal4 | M | 20151.34 | 0.72 |
| fru-M-500231 | 154 | FruMARCM-M002377_seg004 | fru-Gal4 | M | 17320.70 | 0.69 |
| fru-M-200295 | 255 | FruMARCM-M002482_seg001 | fru-Gal4 | M | 20602.62 | 0.75 |
| fru-M-500238 | 338 | FruMARCM-M002572_seg001 | fru-Gal4 | M | 20054.73 | 0.76 |
| fru-M-500160 | 412 | FruMARCM-M001716_seg001 | fru-Gal4 | M | 18340.26 | 0.66 |
| fru-M-300195 | 513 | FruMARCM-M001616_seg001 | fru-Gal4 | M | 19830.58 | 0.72 |
| fru-M-900040 | 599 | FruMARCM-M001791_seg001 | fru-Gal4 | M | 20160.49 | 0.75 |
| fru-M-200228 | 667 | FruMARCM-M001874_seg001 | fru-Gal4 | M | 19919.61 | 0.75 |
| fru-M-000128 | 734 | FruMARCM-M001945_seg002 | fru-Gal4 | M | 19847.47 | 0.75 |
| fru-M-500198 | 764 | FruMARCM-M001978_seg001 | fru-Gal4 | M | 20780.17 | 0.75 |
| fru-M-200241 | 852 | FruMARCM-M002103_seg001 | fru-Gal4 | M | 20496.71 | 0.74 |
| fru-M-700135 | 886 | FruMARCM-M002156_seg001 | fru-Gal4 | M | 20390.72 | 0.71 |
| fru-M-200145 | 967 | FruMARCM-M001144_seg001 | fru-Gal4 | M | 19750.21 | 0.73 |
| fru-M-300122 | 969 | FruMARCM-M001145_seg001 | fru-Gal4 | M | 17909.83 | 0.71 |
| fru-M-500107 | 1019 | FruMARCM-M001203_seg001 | fru-Gal4 | M | 19173.71 | 0.73 |
| fru-M-200177 | 1199 | FruMARCM-M001378_seg002 | fru-Gal4 | M | 19923.17 | 0.77 |
| fru-M-400107 | 1343 | FruMARCM-M001095_seg001 | fru-Gal4 | M | 19732.14 | 0.68 |
| fru-M-500075 | 1389 | FruMARCM-M000893_seg001 | fru-Gal4 | M | 19485.19 | 0.73 |
| fru-M-200098 | 1451 | FruMARCM-M001018_seg002 | fru-Gal4 | M | 18814.44 | 0.72 |
| fru-M-200103 | 1482 | FruMARCM-M001039_seg001 | fru-Gal4 | M | 18156.08 | 0.72 |
| fru-M-300111 | 1494 | FruMARCM-M001047_seg001 | fru-Gal4 | M | 18414.50 | 0.67 |
| fru-M-000022 | 1604 | FruMARCM-M000722_seg002 | fru-Gal4 | M | 19784.59 | 0.73 |
| fru-M-300076 | 1611 | FruMARCM-M000726_seg002 | fru-Gal4 | M | 17876.27 | 0.69 |
| fru-M-200086 | 1620 | FruMARCM-M000741_seg002 | fru-Gal4 | M | 17686.76 | 0.62 |
| fru-M-200092 | 1626 | FruMARCM-M000747_seg001 | fru-Gal4 | M | 19344.77 | 0.73 |
| fru-M-300088 | 1678 | FruMARCM-M000834_seg001 | fru-Gal4 | M | 19759.13 | 0.71 |
| fru-M-100025 | 1771 | FruMARCM-M000288_seg002 | fru-Gal4 | M | 17836.45 | 0.69 |
| fru-M-500037 | 1960 | FruMARCM-M000600_seg003 | fru-Gal4 | M | 17685.67 | 0.70 |
| fru-M-200010 | 2329 | FruMARCM-M000016_seg002 | fru-Gal4 | M | 19692.52 | 0.73 |
| fru-M-300007 | 2360 | FruMARCM-M000053_seg001 | fru-Gal4 | M | 19721.66 | 0.76 |
| fru-F-200025 | 9791 | FruMARCM-F000310_seg001 | fru-Gal4 | F | 17867.24 | 0.71 |