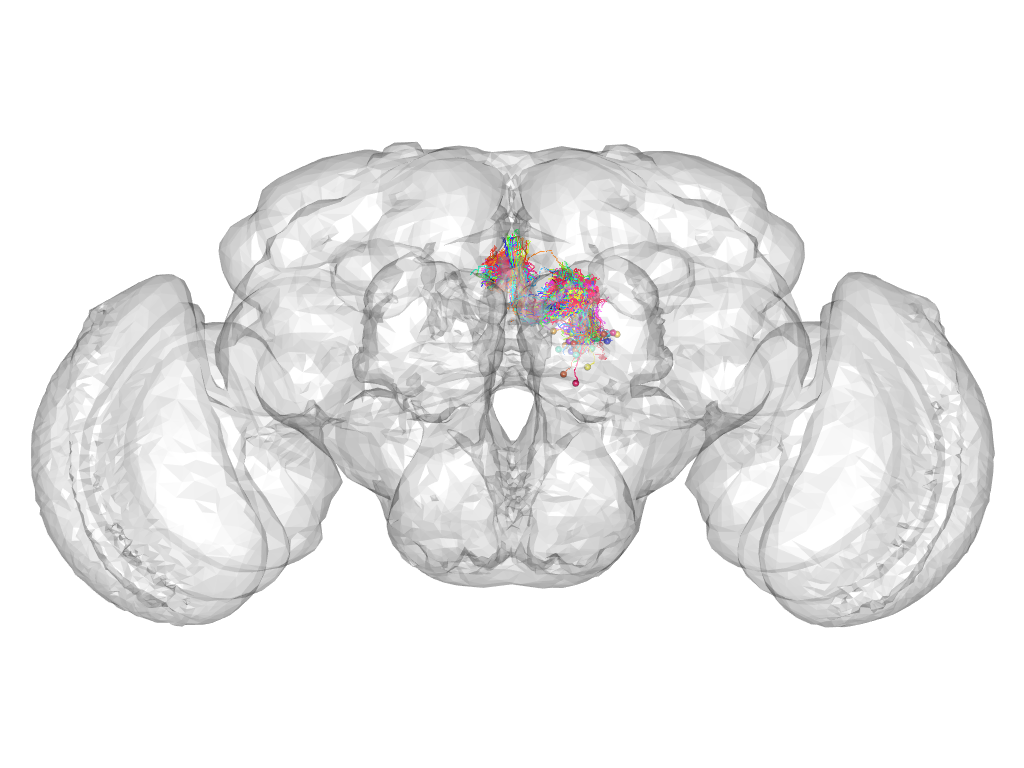
Cluster 769
Part of supercluster XIV
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-500410), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 361 | Cha-F-100099 | XIV | 0.75 |
| 974 | VGlut-F-400120 | XIV | 0.71 |
| 204 | Trh-M-100025 | XIV | 0.70 |
| 192 | TH-M-300027 | XIV | 0.70 |
| 752 | VGlut-F-300443 | XIV | 0.69 |
| 1028 | VGlut-F-000067 | XIV | 0.67 |
| 191 | TH-M-200018 | XIV | 0.67 |
| 139 | npf-M-900000 | XIV | 0.65 |
| 314 | fru-F-600113 | XIV | 0.60 |
| 964 | VGlut-F-200325 | XIV | 0.56 |
| 63 | fru-M-100176 | XIV | 0.53 |
| 1018 | VGlut-F-300036 | XIV | 0.52 |
Cluster composition
This cluster has 118 neurons. The exemplar of this cluster (VGlut-F-500410) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-500410 | 11976 | DvGlutMARCM-F002510_seg001 | VGlut-Gal4 | F | 14202.37 | 1.00 |
| fru-M-400221 | 271 | FruMARCM-M002496_seg002 | fru-Gal4 | M | 10331.65 | 0.70 |
| fru-M-200322 | 330 | FruMARCM-M002552_seg001 | fru-Gal4 | M | 10128.26 | 0.72 |
| fru-M-500173 | 625 | FruMARCM-M001836_seg002 | fru-Gal4 | M | 9978.32 | 0.68 |
| fru-M-800075 | 633 | FruMARCM-M001844_seg001 | fru-Gal4 | M | 10861.01 | 0.76 |
| fru-M-300204 | 634 | FruMARCM-M001845_seg001 | fru-Gal4 | M | 10617.52 | 0.74 |
| fru-M-800042 | 983 | FruMARCM-M001163_seg001 | fru-Gal4 | M | 10473.95 | 0.73 |
| fru-M-700077 | 1018 | FruMARCM-M001202_seg001 | fru-Gal4 | M | 10597.62 | 0.75 |
| fru-M-000036 | 1353 | FruMARCM-M001100_seg002 | fru-Gal4 | M | 10491.32 | 0.74 |
| fru-M-200067 | 1575 | FruMARCM-M000646_seg001 | fru-Gal4 | M | 10505.81 | 0.74 |
| fru-M-200070 | 1578 | FruMARCM-M000647_seg001 | fru-Gal4 | M | 10469.15 | 0.71 |
| fru-M-400034 | 1863 | FruMARCM-M000400_seg001 | fru-Gal4 | M | 10471.99 | 0.73 |
| fru-M-300061 | 1906 | FruMARCM-M000450_seg001 | fru-Gal4 | M | 10263.37 | 0.71 |
| fru-M-100000 | 2350 | FruMARCM-M000043_seg001 | fru-Gal4 | M | 10341.56 | 0.71 |
| VGlut-F-500869 | 5804 | DvGlutMARCM-F004501_seg002 | VGlut-Gal4 | F | 11061.09 | 0.79 |
| VGlut-F-400919 | 5829 | DvGlutMARCM-F004518_seg001 | VGlut-Gal4 | F | 11095.01 | 0.78 |
| VGlut-F-100396 | 5831 | DvGlutMARCM-F004520_seg001 | VGlut-Gal4 | F | 11098.41 | 0.77 |
| VGlut-F-900136 | 5836 | DvGlutMARCM-F004525_seg001 | VGlut-Gal4 | F | 10525.76 | 0.73 |
| VGlut-F-300648 | 5855 | DvGlutMARCM-F004539_seg001 | VGlut-Gal4 | F | 10799.21 | 0.77 |
| VGlut-F-700589 | 5895 | DvGlutMARCM-F004573_seg001 | VGlut-Gal4 | F | 11200.24 | 0.79 |
| VGlut-F-000484 | 6023 | DvGlutMARCM-F002236_seg001 | VGlut-Gal4 | F | 11054.67 | 0.77 |
| VGlut-F-700081 | 6033 | DvGlutMARCM-F002593_seg002 | VGlut-Gal4 | F | 11137.07 | 0.78 |
| VGlut-F-300601 | 6419 | DvGlutMARCM-F004211_seg001 | VGlut-Gal4 | F | 10928.27 | 0.74 |
| VGlut-F-500789 | 6469 | DvGlutMARCM-F004253_seg001 | VGlut-Gal4 | F | 10856.32 | 0.77 |
| VGlut-F-800260 | 6495 | DvGlutMARCM-F004274_seg001 | VGlut-Gal4 | F | 10962.59 | 0.78 |
| VGlut-F-800268 | 6514 | DvGlutMARCM-F004289_seg002 | VGlut-Gal4 | F | 10370.74 | 0.74 |
| VGlut-F-600702 | 6715 | DvGlutMARCM-F003927_seg001 | VGlut-Gal4 | F | 10783.92 | 0.73 |
| VGlut-F-500748 | 6766 | DvGlutMARCM-F003969_seg001 | VGlut-Gal4 | F | 10694.48 | 0.77 |
| VGlut-F-100353 | 6800 | DvGlutMARCM-F003991_seg001 | VGlut-Gal4 | F | 10847.61 | 0.76 |
| VGlut-F-500764 | 6825 | DvGlutMARCM-F004008_seg001 | VGlut-Gal4 | F | 11014.31 | 0.78 |
| VGlut-F-400832 | 6853 | DvGlutMARCM-F004028_seg001 | VGlut-Gal4 | F | 11092.65 | 0.78 |
| VGlut-F-400847 | 6868 | DvGlutMARCM-F004033_seg001 | VGlut-Gal4 | F | 10457.39 | 0.73 |
| VGlut-F-000630 | 6936 | DvGlutMARCM-F004090_seg001 | VGlut-Gal4 | F | 11077.18 | 0.78 |
| VGlut-F-300592 | 6990 | DvGlutMARCM-F004135_seg002 | VGlut-Gal4 | F | 10950.75 | 0.75 |
| VGlut-F-000646 | 7031 | DvGlutMARCM-F004167_seg001 | VGlut-Gal4 | F | 11038.87 | 0.75 |
| VGlut-F-700499 | 7036 | DvGlutMARCM-F004170_seg001 | VGlut-Gal4 | F | 10866.41 | 0.76 |
| VGlut-F-100338 | 7920 | DvGlutMARCM-F003763_seg001 | VGlut-Gal4 | F | 10855.50 | 0.74 |
| VGlut-F-200535 | 7969 | DvGlutMARCM-F003796_seg001 | VGlut-Gal4 | F | 10895.92 | 0.74 |
| VGlut-F-100342 | 8001 | DvGlutMARCM-F003821_seg001 | VGlut-Gal4 | F | 10942.89 | 0.77 |
| VGlut-F-200514 | 8588 | DvGlutMARCM-F003654_seg001 | VGlut-Gal4 | F | 10680.27 | 0.76 |
| VGlut-F-100320 | 8600 | DvGlutMARCM-F003663_seg001 | VGlut-Gal4 | F | 10686.18 | 0.74 |
| VGlut-F-400753 | 8652 | DvGlutMARCM-F003702_seg001 | VGlut-Gal4 | F | 11118.54 | 0.78 |
| VGlut-F-300499 | 8739 | DvGlutMARCM-F003566_seg001 | VGlut-Gal4 | F | 11019.27 | 0.77 |
| VGlut-F-000540 | 8759 | DvGlutMARCM-F003581_seg001 | VGlut-Gal4 | F | 10868.88 | 0.77 |
| VGlut-F-500706 | 8797 | DvGlutMARCM-F003611_seg001 | VGlut-Gal4 | F | 11213.54 | 0.79 |
| VGlut-F-200507 | 8818 | DvGlutMARCM-F003623_seg002 | VGlut-Gal4 | F | 10706.13 | 0.76 |
| VGlut-F-400736 | 8819 | DvGlutMARCM-F003624_seg001 | VGlut-Gal4 | F | 10799.07 | 0.72 |
| VGlut-F-100310 | 8841 | DvGlutMARCM-F003637_seg001 | VGlut-Gal4 | F | 11265.24 | 0.80 |
| VGlut-F-800199 | 8863 | DvGlutMARCM-F003451_seg001 | VGlut-Gal4 | F | 10955.72 | 0.77 |
| VGlut-F-000525 | 8917 | DvGlutMARCM-F003493_seg001 | VGlut-Gal4 | F | 10915.81 | 0.78 |
| VGlut-F-700365 | 8926 | DvGlutMARCM-F003500_seg001 | VGlut-Gal4 | F | 10970.39 | 0.77 |
| VGlut-F-500677 | 8940 | DvGlutMARCM-F003508_seg001 | VGlut-Gal4 | F | 11277.59 | 0.79 |
| VGlut-F-600602 | 9052 | DvGlutMARCM-F003375_seg001 | VGlut-Gal4 | F | 10898.12 | 0.77 |
| VGlut-F-400700 | 9115 | DvGlutMARCM-F003419_seg001 | VGlut-Gal4 | F | 10936.43 | 0.76 |
| VGlut-F-600626 | 9154 | DvGlutMARCM-F003440_seg001 | VGlut-Gal4 | F | 10792.75 | 0.75 |
| VGlut-F-300497 | 9174 | DvGlutMARCM-F003248_seg001 | VGlut-Gal4 | F | 10350.87 | 0.72 |
| VGlut-F-200467 | 9208 | DvGlutMARCM-F003274_seg001 | VGlut-Gal4 | F | 11181.44 | 0.78 |
| VGlut-F-200470 | 9215 | DvGlutMARCM-F003281_seg001 | VGlut-Gal4 | F | 11011.41 | 0.74 |
| VGlut-F-100288 | 9295 | DvGlutMARCM-F003342_seg001 | VGlut-Gal4 | F | 10679.75 | 0.77 |
| VGlut-F-400670 | 9370 | DvGlutMARCM-F003201_seg001 | VGlut-Gal4 | F | 10691.69 | 0.76 |
| VGlut-F-400616 | 10217 | DvGlutMARCM-F002849_seg002 | VGlut-Gal4 | F | 11146.30 | 0.77 |
| VGlut-F-000027 | 10245 | DvGlutMARCM-F197-X3_seg1 | VGlut-Gal4 | F | 10844.36 | 0.78 |
| Gad1-F-600008 | 10584 | GadMARCM-F000101_seg001 | Gad1-Gal4 | F | 11059.63 | 0.77 |
| VGlut-F-400605 | 10884 | DvGlutMARCM-F002791_seg001 | VGlut-Gal4 | F | 11072.69 | 0.78 |
| VGlut-F-700199 | 10935 | DvGlutMARCM-F002829_seg001 | VGlut-Gal4 | F | 11102.63 | 0.77 |
| VGlut-F-800083 | 10962 | DvGlutMARCM-F002856_seg002 | VGlut-Gal4 | F | 11147.36 | 0.79 |
| VGlut-F-800086 | 10974 | DvGlutMARCM-F002864_seg001 | VGlut-Gal4 | F | 11008.22 | 0.78 |
| VGlut-F-800087 | 10975 | DvGlutMARCM-F002865_seg001 | VGlut-Gal4 | F | 10478.82 | 0.73 |
| VGlut-F-500478 | 10979 | DvGlutMARCM-F002869_seg001 | VGlut-Gal4 | F | 10972.90 | 0.77 |
| VGlut-F-200432 | 11004 | DvGlutMARCM-F002890_seg001 | VGlut-Gal4 | F | 10693.11 | 0.75 |
| VGlut-F-500482 | 11010 | DvGlutMARCM-F002895_seg001 | VGlut-Gal4 | F | 10914.62 | 0.77 |
| VGlut-F-100263 | 11041 | DvGlutMARCM-F002920_seg001 | VGlut-Gal4 | F | 10774.59 | 0.75 |
| VGlut-F-600467 | 11076 | DvGlutMARCM-F002954_seg001 | VGlut-Gal4 | F | 10880.72 | 0.73 |
| VGlut-F-700229 | 11094 | DvGlutMARCM-F002967_seg001 | VGlut-Gal4 | F | 10671.13 | 0.74 |
| VGlut-F-900066 | 11104 | DvGlutMARCM-F002973_seg001 | VGlut-Gal4 | F | 11195.84 | 0.79 |
| VGlut-F-600483 | 11165 | DvGlutMARCM-F003014_seg001 | VGlut-Gal4 | F | 11100.56 | 0.78 |
| fru-X-400004 | 11555 | FruMARCM-X000025_seg001 | fru-Gal4 | F | 10124.51 | 0.72 |
| VGlut-F-100249 | 11689 | DvGlutMARCM-F002264_seg001 | VGlut-Gal4 | F | 11013.57 | 0.77 |
| VGlut-F-100254 | 11694 | DvGlutMARCM-F002268_seg001 | VGlut-Gal4 | F | 10565.86 | 0.70 |
| VGlut-F-100255 | 11695 | DvGlutMARCM-F002269_seg001 | VGlut-Gal4 | F | 10913.65 | 0.76 |
| VGlut-F-300408 | 11931 | DvGlutMARCM-F002466_seg001 | VGlut-Gal4 | F | 10801.79 | 0.76 |
| VGlut-F-900032 | 11941 | DvGlutMARCM-F002475_seg002 | VGlut-Gal4 | F | 10887.12 | 0.78 |
| VGlut-F-100260 | 11959 | DvGlutMARCM-F002492_seg001 | VGlut-Gal4 | F | 10924.98 | 0.77 |
| VGlut-F-500416 | 12007 | DvGlutMARCM-F002529_seg001 | VGlut-Gal4 | F | 10894.94 | 0.78 |
| VGlut-F-600312 | 12041 | DvGlutMARCM-F002555_seg002 | VGlut-Gal4 | F | 10842.96 | 0.77 |
| VGlut-F-700074 | 12081 | DvGlutMARCM-F002590_seg001 | VGlut-Gal4 | F | 10282.20 | 0.73 |
| VGlut-F-800049 | 12092 | DvGlutMARCM-F002597_seg001 | VGlut-Gal4 | F | 11053.49 | 0.78 |
| VGlut-F-500465 | 12233 | DvGlutMARCM-F002697_seg001 | VGlut-Gal4 | F | 11038.68 | 0.75 |
| VGlut-F-700108 | 12248 | DvGlutMARCM-F002711_seg001 | VGlut-Gal4 | F | 11051.30 | 0.77 |
| VGlut-F-400582 | 12288 | DvGlutMARCM-F002743_seg001 | VGlut-Gal4 | F | 11140.10 | 0.79 |
| VGlut-F-400584 | 12290 | DvGlutMARCM-F002745_seg001 | VGlut-Gal4 | F | 10997.69 | 0.78 |
| VGlut-F-200338 | 12804 | DvGlutMARCM-F1849_seg1 | VGlut-Gal4 | F | 9352.47 | 0.73 |
| VGlut-F-500283 | 12810 | DvGlutMARCM-F1858_seg1 | VGlut-Gal4 | F | 10879.49 | 0.77 |
| VGlut-F-400463 | 12892 | DvGlutMARCM-F1935_seg1 | VGlut-Gal4 | F | 10940.23 | 0.75 |
| VGlut-F-700041 | 13063 | DvGlutMARCM-F2087_seg1 | VGlut-Gal4 | F | 10531.15 | 0.75 |
| VGlut-F-300191 | 14342 | DvGlutMARCM-F1026_seg1 | VGlut-Gal4 | F | 10930.55 | 0.78 |
| VGlut-F-000300 | 14414 | DvGlutMARCM-F1094_seg1 | VGlut-Gal4 | F | 11169.35 | 0.77 |
| VGlut-F-400255 | 14625 | DvGlutMARCM-F883_seg1 | VGlut-Gal4 | F | 11091.52 | 0.77 |
| VGlut-F-400279 | 14676 | DvGlutMARCM-F937_seg1 | VGlut-Gal4 | F | 10658.93 | 0.74 |
| VGlut-F-200227 | 14829 | DvGlutMARCM-F1171_seg1 | VGlut-Gal4 | F | 10862.06 | 0.75 |
| VGlut-F-200243 | 14882 | DvGlutMARCM-F1226_seg1 | VGlut-Gal4 | F | 11138.46 | 0.77 |
| VGlut-F-800022 | 14914 | DvGlutMARCM-F1257_seg1 | VGlut-Gal4 | F | 10909.38 | 0.75 |
| VGlut-F-400349 | 14929 | DvGlutMARCM-F1271_seg1 | VGlut-Gal4 | F | 10674.58 | 0.75 |
| VGlut-F-100138 | 14970 | DvGlutMARCM-F1308_seg1 | VGlut-Gal4 | F | 11156.25 | 0.79 |
| VGlut-F-400383 | 15162 | DvGlutMARCM-F1494_seg1 | VGlut-Gal4 | F | 10736.51 | 0.73 |
| VGlut-F-300271 | 15180 | DvGlutMARCM-F1511_seg1 | VGlut-Gal4 | F | 10625.77 | 0.77 |
| VGlut-F-300273 | 15183 | DvGlutMARCM-F1514_seg1 | VGlut-Gal4 | F | 10623.78 | 0.76 |
| VGlut-F-400391 | 15207 | DvGlutMARCM-F1535_seg1 | VGlut-Gal4 | F | 10346.71 | 0.74 |
| VGlut-F-300287 | 15234 | DvGlutMARCM-F1557_seg1 | VGlut-Gal4 | F | 10843.08 | 0.76 |
| VGlut-F-600089 | 15374 | DvGlutMARCM-F1727_seg1 | VGlut-Gal4 | F | 10535.75 | 0.74 |
| VGlut-F-700025 | 15409 | DvGlutMARCM-F1767_seg1 | VGlut-Gal4 | F | 11065.05 | 0.78 |
| VGlut-F-000006 | 15612 | DvGlutMARCM-F028_seg1 | VGlut-Gal4 | F | 10657.39 | 0.72 |
| VGlut-F-300044 | 15920 | DvGlutMARCM-F315_seg1 | VGlut-Gal4 | F | 10466.61 | 0.73 |
| VGlut-F-400074 | 15971 | DvGlutMARCM-F362_seg1 | VGlut-Gal4 | F | 11474.85 | 0.73 |
| VGlut-F-000073 | 16004 | DvGlutMARCM-F393_seg1 | VGlut-Gal4 | F | 11199.20 | 0.76 |
| VGlut-F-100034 | 16016 | DvGlutMARCM-F405_seg1 | VGlut-Gal4 | F | 11017.58 | 0.77 |
| VGlut-F-000097 | 16080 | DvGlutMARCM-F464_seg1 | VGlut-Gal4 | F | 10571.90 | 0.71 |
| VGlut-F-200107 | 16209 | DvGlutMARCM-F588_seg1 | VGlut-Gal4 | F | 10644.17 | 0.75 |