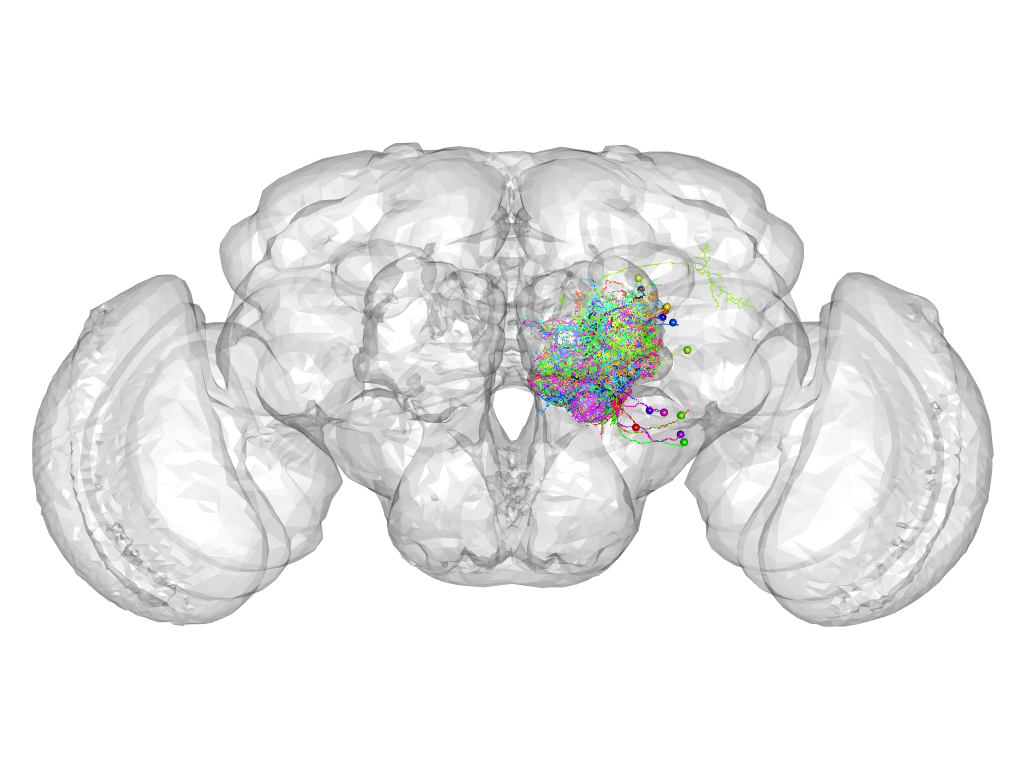
Cluster 748
Part of supercluster XII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (5-HT1B-F-500012), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 806 | Trh-F-100085 | XII | 0.75 |
| 144 | Trh-M-400113 | XII | 0.75 |
| 662 | Tdc2-F-400026 | XIII | 0.67 |
| 400 | VGlut-F-500072 | XII | 0.61 |
| 545 | VGlut-F-700278 | XII | 0.59 |
| 932 | VGlut-F-100102 | XII | 0.59 |
| 653 | Trh-F-000043 | XIX | 0.59 |
| 172 | Trh-M-100092 | XII | 0.56 |
| 513 | Cha-F-300036 | XII | 0.56 |
| 926 | VGlut-F-500134 | XII | 0.53 |
Cluster composition
This cluster has 22 neurons. The exemplar of this cluster (5-HT1B-F-500012) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| 5-HT1B-F-500012 | 11617 | 5HT1bMARCM-F000012_seg001 | 5-HT1B-Gal4 | F | 24429.90 | 1.00 |
| fru-M-200038 | 2448 | FruMARCM-M000209_seg004 | fru-Gal4 | M | 7679.92 | 0.50 |
| 5-HT1B-M-500001 | 2488 | 5HT1bMARCM-M000034_seg001 | 5-HT1B-Gal4 | M | 18019.71 | 0.70 |
| 5-HT1B-M-500003 | 2490 | 5HT1bMARCM-M000036_seg001 | 5-HT1B-Gal4 | M | 18521.23 | 0.75 |
| 5-HT1B-M-500008 | 2526 | 5HT1bMARCM-M000067_seg001 | 5-HT1B-Gal4 | M | 18525.62 | 0.74 |
| Trh-M-300022 | 3519 | TPHMARCM-399M_seg1 | Trh-Gal4 | M | 17910.20 | 0.73 |
| Cha-F-000239 | 5100 | ChaMARCM-F001159_seg001 | Cha-Gal4 | F | 17068.37 | 0.58 |
| VGlut-F-300398 | 6119 | DvGlutMARCM-F2196_seg2 | VGlut-Gal4 | F | 15821.20 | 0.57 |
| VGlut-F-300106 | 10757 | DVGlutMARCM-F-0513-01-F636_seg1 | VGlut-Gal4 | F | 16308.97 | 0.60 |
| 5-HT1B-F-500014 | 11194 | 5HT1bMARCM-F000025_seg001 | 5-HT1B-Gal4 | F | 18532.06 | 0.72 |
| 5-HT1B-F-500015 | 11195 | 5HT1bMARCM-F000025_seg002 | 5-HT1B-Gal4 | F | 18092.12 | 0.71 |
| 5-HT1B-F-500018 | 11198 | 5HT1bMARCM-F000028_seg001 | 5-HT1B-Gal4 | F | 18235.05 | 0.72 |
| 5-HT1B-F-500020 | 11200 | 5HT1bMARCM-F000030_seg001 | 5-HT1B-Gal4 | F | 17431.66 | 0.68 |
| 5-HT1B-F-500000 | 11605 | 5HT1bMARCM-F000002_seg001 | 5-HT1B-Gal4 | F | 18611.77 | 0.73 |
| 5-HT1B-F-500004 | 11609 | 5HT1bMARCM-F000006_seg001 | 5-HT1B-Gal4 | F | 17967.55 | 0.71 |
| 5-HT1B-F-500010 | 11615 | 5HT1bMARCM-F000011_seg001 | 5-HT1B-Gal4 | F | 18249.69 | 0.73 |
| VGlut-F-500518 | 11710 | DvGlutMARCM-F002283_seg001 | VGlut-Gal4 | F | 12388.33 | 0.58 |
| VGlut-F-300395 | 13159 | DvGlutMARCM-F2195_seg1 | VGlut-Gal4 | F | 16253.52 | 0.59 |
| VGlut-F-200264 | 14982 | DvGlutMARCM-F1321_seg1 | VGlut-Gal4 | F | 14317.34 | 0.59 |
| VGlut-F-300286 | 15233 | DvGlutMARCM-F1556_seg1 | VGlut-Gal4 | F | 11135.47 | 0.56 |
| VGlut-F-200023 | 15629 | DvGlutMARCM-F044_seg1 | VGlut-Gal4 | F | 15889.10 | 0.59 |
| VGlut-F-200025 | 15631 | DvGlutMARCM-F045-X2_seg2 | VGlut-Gal4 | F | 16030.06 | 0.58 |