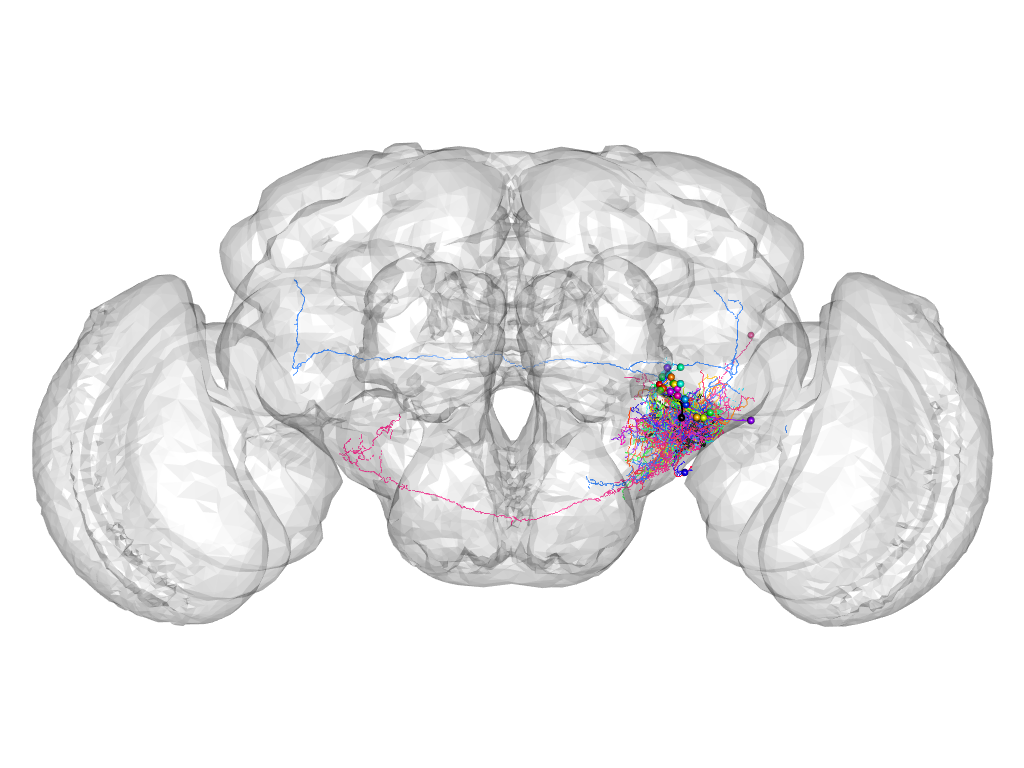
Cluster 734
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-800001), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 159 | fru-M-700004 | VIII | 0.70 |
| 962 | VGlut-F-600088 | X | 0.62 |
| 652 | Tdc2-F-200010 | VIII | 0.59 |
| 988 | VGlut-F-200005 | X | 0.58 |
| 27 | fru-M-300302 | VIII | 0.51 |
Cluster composition
This cluster has 26 neurons. The exemplar of this cluster (fru-F-800001) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-800001 | 11442 | FruMARCM-F000063_seg001 | fru-Gal4 | F | 17539.41 | 1.00 |
| fru-M-200286 | 160 | FruMARCM-M002382_seg001 | fru-Gal4 | M | 12734.74 | 0.70 |
| fru-M-700186 | 370 | FruMARCM-M002605_seg001 | fru-Gal4 | M | 12532.71 | 0.67 |
| fru-M-000076 | 482 | FruMARCM-M001583_seg001 | fru-Gal4 | M | 12586.27 | 0.72 |
| fru-M-600075 | 579 | FruMARCM-M001767_seg001 | fru-Gal4 | M | 12591.06 | 0.69 |
| fru-M-400165 | 596 | FruMARCM-M001788_seg001 | fru-Gal4 | M | 12659.97 | 0.72 |
| fru-M-700100 | 622 | FruMARCM-M001827_seg001 | fru-Gal4 | M | 11750.16 | 0.69 |
| fru-M-400170 | 671 | FruMARCM-M001878_seg001 | fru-Gal4 | M | 12974.43 | 0.72 |
| fru-M-600066 | 1273 | FruMARCM-M001462_seg001 | fru-Gal4 | M | 12119.02 | 0.73 |
| fru-M-500100 | 1333 | FruMARCM-M001090_seg002 | fru-Gal4 | M | 12584.36 | 0.72 |
| fru-M-400085 | 1460 | FruMARCM-M001023_seg001 | fru-Gal4 | M | 12040.74 | 0.61 |
| fru-M-200115 | 1513 | FruMARCM-M001061_seg001 | fru-Gal4 | M | 12322.06 | 0.71 |
| fru-M-800016 | 1764 | FruMARCM-M000252_seg001 | fru-Gal4 | M | 12147.76 | 0.70 |
| fru-M-700022 | 1788 | FruMARCM-M000317_seg001 | fru-Gal4 | M | 12049.82 | 0.69 |
| fru-M-700036 | 1892 | FruMARCM-M000425_seg003 | fru-Gal4 | M | 11842.28 | 0.63 |
| fru-M-700037 | 1893 | FruMARCM-M000425_seg004 | fru-Gal4 | M | 12024.35 | 0.71 |
| Cha-F-100247 | 4139 | ChaMARCM-F001257_seg001 | Cha-Gal4 | F | 12117.61 | 0.42 |
| fru-F-600090 | 4502 | FruMARCM-F001604_seg002 | fru-Gal4 | F | 12759.04 | 0.73 |
| Cha-F-000131 | 5354 | ChaMARCM-F000724_seg001 | Cha-Gal4 | F | 8072.85 | 0.46 |
| fru-F-700135 | 6297 | FruMARCM-F001394_seg001 | fru-Gal4 | F | 12199.80 | 0.71 |
| fru-F-700141 | 6312 | FruMARCM-F001432_seg001 | fru-Gal4 | F | 12079.13 | 0.59 |
| fru-F-700066 | 7273 | FruMARCM-F000910_seg001 | fru-Gal4 | F | 12815.57 | 0.75 |
| fru-F-700084 | 7296 | FruMARCM-F000924_seg001 | fru-Gal4 | F | 12468.57 | 0.72 |
| fru-F-700061 | 7788 | FruMARCM-F000685_seg001 | fru-Gal4 | F | 11681.93 | 0.66 |
| VGlut-F-000526 | 8956 | DvGlutMARCM-F003518_seg001 | VGlut-Gal4 | F | 12512.26 | 0.49 |
| Trh-F-500056 | 14182 | TPHMARCM-621F_seg1 | Trh-Gal4 | F | 9353.67 | 0.53 |