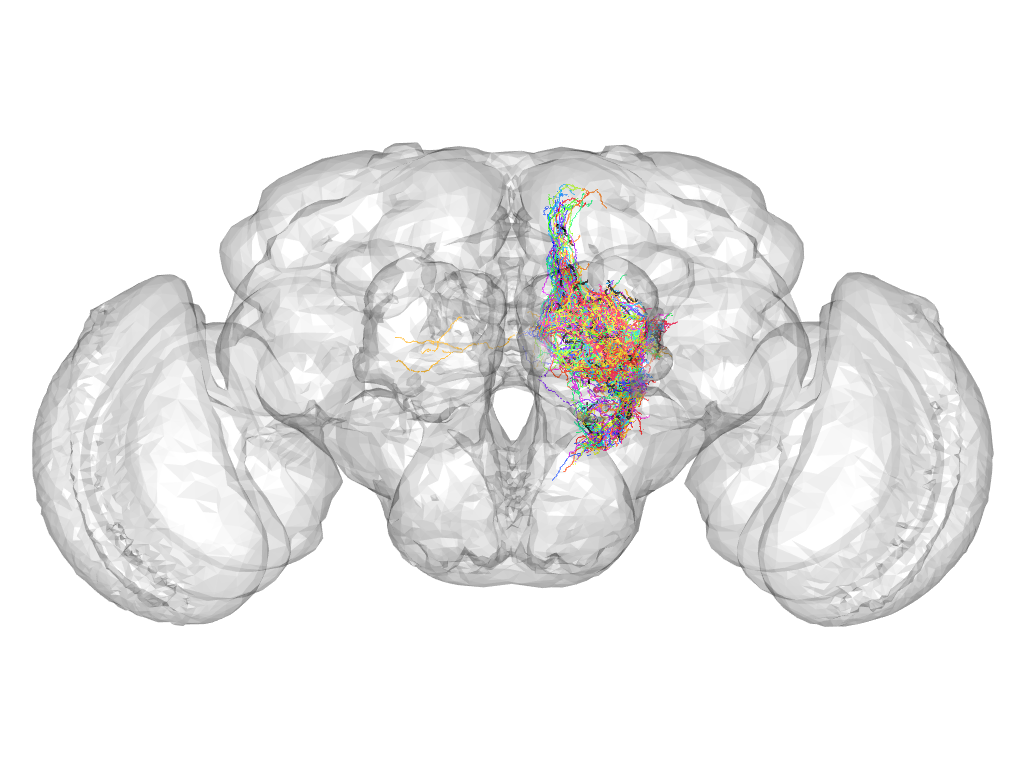
Cluster 73
Part of supercluster XII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-100191), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 129 | fru-M-500019 | XII | 0.68 |
| 48 | fru-M-500155 | XII | 0.65 |
| 857 | TH-F-300062 | XII | 0.63 |
| 31 | fru-M-000206 | XII | 0.60 |
| 215 | fru-F-100075 | XII | 0.60 |
| 1046 | VGlut-F-000154 | XII | 0.57 |
| 240 | Gad1-F-600092 | XII | 0.55 |
| 220 | fru-F-200158 | XII | 0.54 |
| 863 | TH-F-000022 | XII | 0.53 |
| 273 | Cha-F-000338 | XII | 0.51 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (fru-M-100191) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-100191 | 845 | FruMARCM-M002098_seg001 | fru-Gal4 | M | 54144.40 | 1.00 |
| fru-M-200266 | 1 | FruMARCM-M002262_seg001 | fru-Gal4 | M | 38838.61 | 0.69 |
| fru-M-900052 | 106 | FruMARCM-M002305_seg001 | fru-Gal4 | M | 37939.52 | 0.72 |
| fru-M-200291 | 233 | FruMARCM-M002446_seg001 | fru-Gal4 | M | 39012.06 | 0.72 |
| fru-M-200321 | 329 | FruMARCM-M002551_seg001 | fru-Gal4 | M | 38530.05 | 0.67 |
| fru-M-000204 | 333 | FruMARCM-M002555_seg001 | fru-Gal4 | M | 36132.96 | 0.70 |
| fru-M-700088 | 465 | FruMARCM-M001570_seg001 | fru-Gal4 | M | 34962.46 | 0.69 |
| fru-M-800081 | 717 | FruMARCM-M001921_seg001 | fru-Gal4 | M | 38609.08 | 0.72 |
| fru-M-100119 | 1138 | FruMARCM-M001331_seg001 | fru-Gal4 | M | 37292.88 | 0.71 |
| fru-M-100124 | 1158 | FruMARCM-M001347_seg002 | fru-Gal4 | M | 36914.94 | 0.71 |
| fru-M-300166 | 1290 | FruMARCM-M001477_seg002 | fru-Gal4 | M | 37552.49 | 0.73 |
| fru-M-400088 | 1463 | FruMARCM-M001024_seg001 | fru-Gal4 | M | 37940.34 | 0.71 |
| fru-M-200134 | 1548 | FruMARCM-M001081_seg001 | fru-Gal4 | M | 36374.45 | 0.67 |
| fru-M-000023 | 1605 | FruMARCM-M000723_seg001 | fru-Gal4 | M | 36539.68 | 0.72 |
| fru-M-500053 | 1658 | FruMARCM-M000805_seg003 | fru-Gal4 | M | 34150.72 | 0.68 |
| fru-M-400052 | 1683 | FruMARCM-M000837_seg001 | fru-Gal4 | M | 37183.74 | 0.71 |
| fru-M-300089 | 1695 | FruMARCM-M000848_seg001 | fru-Gal4 | M | 34758.72 | 0.68 |
| fru-M-100041 | 1880 | FruMARCM-M000420_seg002 | fru-Gal4 | M | 38078.07 | 0.72 |
| fru-M-300055 | 1900 | FruMARCM-M000436_seg002 | fru-Gal4 | M | 35954.12 | 0.69 |
| fru-M-200012 | 2331 | FruMARCM-M000018_seg001 | fru-Gal4 | M | 36802.80 | 0.68 |
| fru-M-900020 | 2439 | FruMARCM-M000155_seg001 | fru-Gal4 | M | 39019.28 | 0.74 |
| fru-M-200033 | 2443 | FruMARCM-M000207_seg001 | fru-Gal4 | M | 37231.90 | 0.69 |
| fru-F-200140 | 4689 | FruMARCM-F001923_seg001 | fru-Gal4 | F | 37893.08 | 0.72 |
| fru-F-700131 | 6293 | FruMARCM-F001391_seg001 | fru-Gal4 | F | 37154.53 | 0.70 |
| fru-F-300075 | 6393 | FruMARCM-F001530_seg001 | fru-Gal4 | F | 38901.03 | 0.73 |
| fru-F-100032 | 7799 | FruMARCM-F000699_seg001 | fru-Gal4 | F | 33021.99 | 0.66 |
| fru-F-500147 | 7847 | FruMARCM-F000756_seg001 | fru-Gal4 | F | 36574.63 | 0.70 |
| fru-F-600050 | 7902 | FruMARCM-F000845_seg003 | fru-Gal4 | F | 33723.37 | 0.66 |