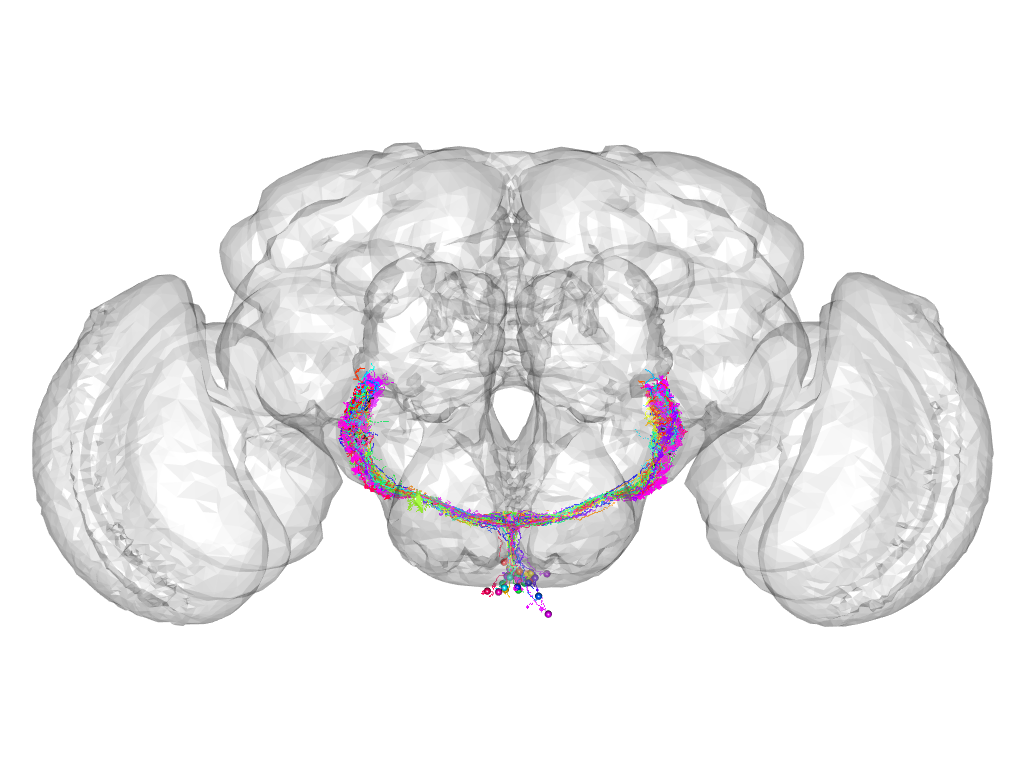
Cluster 681
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Gad1-F-500002), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 652 | Tdc2-F-200010 | VIII | 0.48 |
| 146 | Trh-M-200088 | VIII | 0.48 |
| 177 | Trh-M-300109 | VIII | 0.12 |
| 847 | TH-F-000004 | IX | 0.10 |
| 960 | VGlut-F-500219 | VIII | 0.03 |
Cluster composition
This cluster has 34 neurons. The exemplar of this cluster (Gad1-F-500002) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Gad1-F-500002 | 10713 | GadMARCM-F000043_seg003 | Gad1-Gal4 | F | 19452.80 | 1.00 |
| Gad1-F-200124 | 3970 | GadMARCM-F000595_seg002 | Gad1-Gal4 | F | 14479.23 | 0.74 |
| Gad1-F-200126 | 3972 | GadMARCM-F000596_seg002 | Gad1-Gal4 | F | 15231.00 | 0.76 |
| Cha-F-400204 | 5634 | ChaMARCM-F000912_seg002 | Cha-Gal4 | F | 14550.15 | 0.75 |
| Gad1-F-500041 | 6008 | GadMARCM-F000171_seg002 | Gad1-Gal4 | F | 15124.57 | 0.75 |
| Gad1-F-200053 | 6011 | GadMARCM-F000194_seg003 | Gad1-Gal4 | F | 15532.27 | 0.77 |
| VGlut-F-300612 | 6440 | DvGlutMARCM-F004227_seg003 | VGlut-Gal4 | F | 12560.57 | 0.69 |
| Cha-F-700097 | 7230 | ChaMARCM-F000404_seg003 | Cha-Gal4 | F | 15171.56 | 0.75 |
| Cha-F-700127 | 7655 | ChaMARCM-F000486_seg001 | Cha-Gal4 | F | 14514.23 | 0.75 |
| VGlut-F-600608 | 9136 | DvGlutMARCM-F003428_seg002 | VGlut-Gal4 | F | 8760.72 | 0.53 |
| VGlut-F-600582 | 9260 | DvGlutMARCM-F003318_seg002 | VGlut-Gal4 | F | 14652.72 | 0.75 |
| Gad1-F-000039 | 9512 | GadMARCM-F000401_seg004 | Gad1-Gal4 | F | 14821.20 | 0.77 |
| Gad1-F-800062 | 9698 | GadMARCM-F000543_seg003 | Gad1-Gal4 | F | 14466.77 | 0.76 |
| Gad1-F-500024 | 10508 | GadMARCM-F000155_seg003 | Gad1-Gal4 | F | 12699.11 | 0.70 |
| Gad1-F-800014 | 10689 | GadMARCM-F000189_seg002 | Gad1-Gal4 | F | 14467.39 | 0.74 |
| VGlut-F-900063 | 11090 | DvGlutMARCM-F002963_seg001 | VGlut-Gal4 | F | 14383.01 | 0.75 |
| Gad1-F-100027 | 11273 | GadMARCM-F000248_seg002 | Gad1-Gal4 | F | 15400.48 | 0.77 |
| Gad1-F-200084 | 11330 | GadMARCM-F000299_seg003 | Gad1-Gal4 | F | 15215.58 | 0.77 |
| Gad1-F-300088 | 11386 | GadMARCM-F000339_seg002 | Gad1-Gal4 | F | 14945.12 | 0.75 |
| VGlut-F-200368 | 13008 | DvGlutMARCM-F2036_seg1 | VGlut-Gal4 | F | 14265.83 | 0.73 |
| VGlut-F-600127 | 13107 | DvGlutMARCM-F2132_seg1 | VGlut-Gal4 | F | 10379.31 | 0.67 |
| VGlut-F-200303 | 15323 | DvGlutMARCM-F1663_seg1 | VGlut-Gal4 | F | 13776.03 | 0.72 |
| VGlut-F-300320 | 15344 | DvGlutMARCM-F1691_seg1 | VGlut-Gal4 | F | 14008.07 | 0.74 |
| VGlut-F-500231 | 15347 | DvGlutMARCM-F1696_seg1 | VGlut-Gal4 | F | 13859.53 | 0.70 |
| VGlut-F-300097 | 15442 | DvGlutMARCM-F601_seg1 | VGlut-Gal4 | F | 14018.87 | 0.75 |
| VGlut-F-400031 | 15725 | DvGlutMARCM-F137_seg1 | VGlut-Gal4 | F | 14473.10 | 0.73 |
| VGlut-F-800005 | 15747 | DvGlutMARCM-F156_seg1 | VGlut-Gal4 | F | 10516.30 | 0.58 |
| VGlut-F-300020 | 15798 | DvGlutMARCM-F209_seg1 | VGlut-Gal4 | F | 14859.61 | 0.75 |
| VGlut-F-300060 | 15937 | DvGlutMARCM-F332_seg1 | VGlut-Gal4 | F | 13517.70 | 0.65 |
| VGlut-F-000054 | 15972 | DvGlutMARCM-F363_seg1 | VGlut-Gal4 | F | 14756.64 | 0.71 |
| VGlut-F-000068 | 15996 | DvGlutMARCM-F384_seg1 | VGlut-Gal4 | F | 14688.75 | 0.75 |
| VGlut-F-500087 | 16058 | DvGlutMARCM-F445_seg1 | VGlut-Gal4 | F | 14884.16 | 0.77 |
| VGlut-F-300078 | 16123 | DvGlutMARCM-F507_seg1 | VGlut-Gal4 | F | 9690.84 | 0.55 |
| VGlut-F-000136 | 16153 | DvGlutMARCM-F536_seg1 | VGlut-Gal4 | F | 12258.58 | 0.67 |