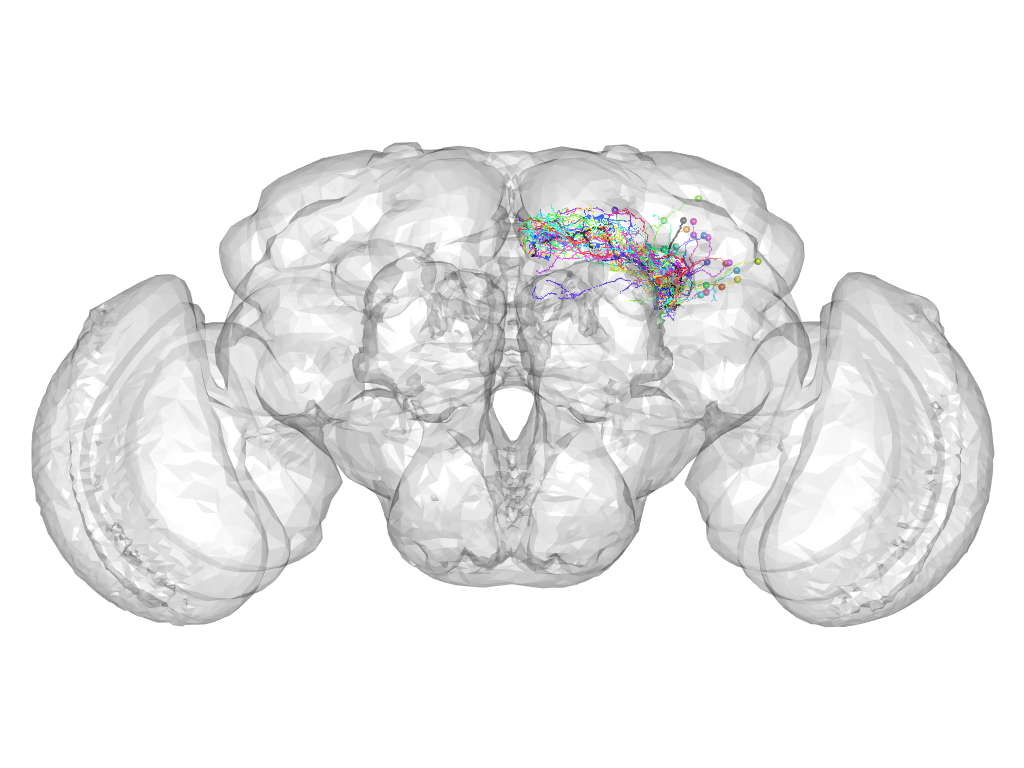
Cluster 51
Part of supercluster XV
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-000088), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 1048 | VGlut-F-000167 | XV | 0.49 |
| 739 | fru-F-200006 | XV | 0.48 |
| 743 | fru-F-000002 | XV | 0.47 |
| 88 | fru-M-500118 | XVI | 0.42 |
| 277 | Cha-F-100326 | XV | 0.39 |
Cluster composition
This cluster has 25 neurons. The exemplar of this cluster (fru-M-000088) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-000088 | 557 | FruMARCM-M001729_seg001 | fru-Gal4 | M | 8348.31 | 1.00 |
| fru-M-500152 | 448 | FruMARCM-M001559_seg001 | fru-Gal4 | M | 4337.49 | 0.50 |
| fru-M-200130 | 1542 | FruMARCM-M001075_seg002 | fru-Gal4 | M | 4508.82 | 0.56 |
| fru-M-000006 | 1930 | FruMARCM-M000524_seg001 | fru-Gal4 | M | 3969.59 | 0.51 |
| fru-M-200015 | 2334 | FruMARCM-M000029_seg002 | fru-Gal4 | M | 2890.58 | 0.53 |
| fru-M-200041 | 2453 | FruMARCM-M000212_seg002 | fru-Gal4 | M | 4489.74 | 0.52 |
| fru-M-300030 | 2462 | FruMARCM-M000220_seg001 | fru-Gal4 | M | 3897.93 | 0.50 |
| 5-HT1B-M-000008 | 2535 | 5HT1bMARCM-M000074_seg001 | 5-HT1B-Gal4 | M | 4218.00 | 0.50 |
| Cha-F-100167 | 4911 | ChaMARCM-F001023_seg001 | Cha-Gal4 | F | 4700.10 | 0.54 |
| Cha-F-100184 | 4933 | ChaMARCM-F001039_seg001 | Cha-Gal4 | F | 4290.70 | 0.59 |
| fru-F-700083 | 7295 | FruMARCM-F000923_seg003 | fru-Gal4 | F | 4073.47 | 0.51 |
| fru-F-500153 | 7305 | FruMARCM-F000945_seg003 | fru-Gal4 | F | 4210.54 | 0.50 |
| Cha-F-300090 | 8319 | ChaMARCM-F000280_seg001 | Cha-Gal4 | F | 3917.65 | 0.51 |
| Gad1-F-200070 | 8403 | GadMARCM-F000260_seg001 | Gad1-Gal4 | F | 4858.34 | 0.63 |
| fru-F-400037 | 9742 | FruMARCM-F000258_seg001 | fru-Gal4 | F | 3995.33 | 0.48 |
| Gad1-F-400018 | 10567 | GadMARCM-F000090_seg001 | Gad1-Gal4 | F | 3617.85 | 0.48 |
| VGlut-F-000478 | 11110 | DvGlutMARCM-F002976_seg001 | VGlut-Gal4 | F | 4261.54 | 0.51 |
| Gad1-F-100021 | 11263 | GadMARCM-F000241_seg001 | Gad1-Gal4 | F | 4022.89 | 0.53 |
| Gad1-F-800018 | 11308 | GadMARCM-F000281_seg001 | Gad1-Gal4 | F | 2234.40 | 0.23 |
| Gad1-F-600039 | 11361 | GadMARCM-F000319_seg001 | Gad1-Gal4 | F | 4800.97 | 0.56 |
| fru-F-700023 | 11532 | FruMARCM-F000190_seg001 | fru-Gal4 | F | 3901.43 | 0.51 |
| fru-X-400003 | 11554 | FruMARCM-X000024_seg002 | fru-Gal4 | F | 4438.05 | 0.52 |
| 5-HT1B-F-000003 | 11621 | 5HT1bMARCM-F000016_seg001 | 5-HT1B-Gal4 | F | 4181.39 | 0.55 |
| 5-HT1B-F-200001 | 11631 | 5HT1bMARCM-F000022_seg002 | 5-HT1B-Gal4 | F | 3284.47 | 0.50 |
| VGlut-F-000308 | 14783 | DvGlutMARCM-F1132_seg1 | VGlut-Gal4 | F | 2990.46 | 0.44 |