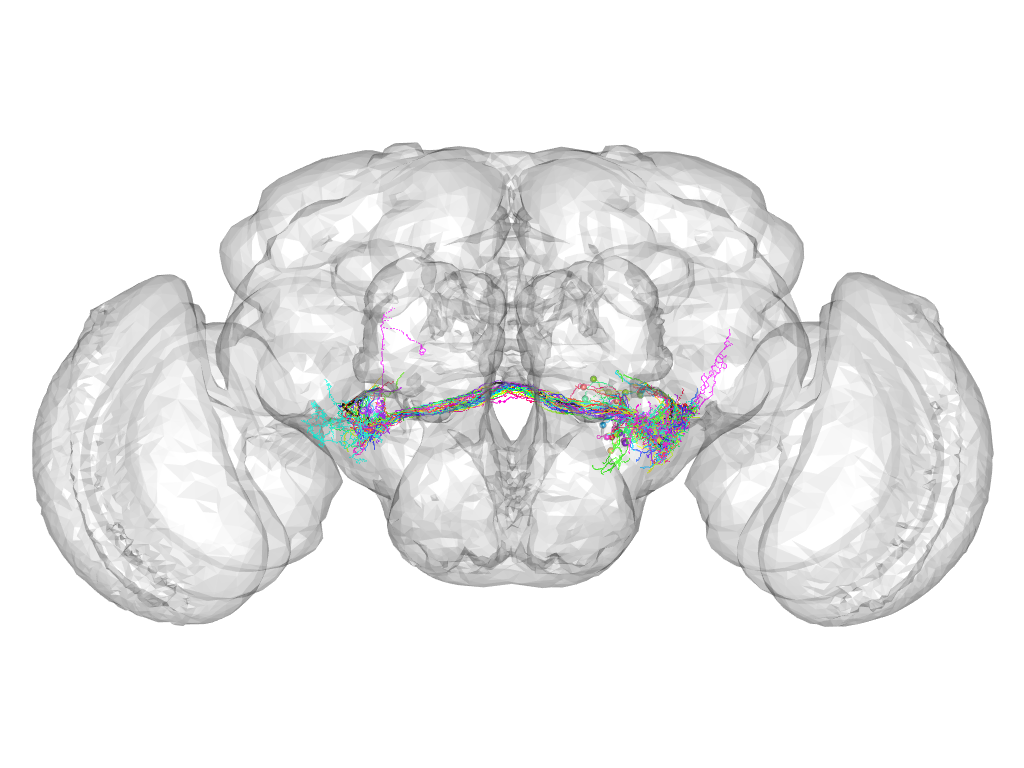
Cluster 47
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-300198), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 320 | Cha-F-400233 | VIII | 0.67 |
| 27 | fru-M-300302 | VIII | 0.61 |
| 902 | VGlut-F-400300 | VIII | 0.61 |
| 459 | VGlut-F-500779 | VIII | 0.61 |
| 410 | fru-F-400147 | VIII | 0.53 |
| 309 | fru-F-700160 | VIII | 0.52 |
Cluster composition
This cluster has 30 neurons. The exemplar of this cluster (fru-M-300198) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-300198 | 516 | FruMARCM-M001629_seg001 | fru-Gal4 | M | 9851.68 | 1.00 |
| fru-M-400203 | 65 | FruMARCM-M002259_seg001 | fru-Gal4 | M | 7472.28 | 0.73 |
| fru-M-000172 | 96 | FruMARCM-M002295_seg001 | fru-Gal4 | M | 6992.70 | 0.70 |
| fru-M-300174 | 427 | FruMARCM-M001545_seg001 | fru-Gal4 | M | 9470.87 | 0.97 |
| fru-M-300187 | 485 | FruMARCM-M001585_seg001 | fru-Gal4 | M | 7258.57 | 0.74 |
| fru-M-400174 | 696 | FruMARCM-M001899_seg001 | fru-Gal4 | M | 7524.16 | 0.74 |
| fru-M-400183 | 759 | FruMARCM-M001972_seg001 | fru-Gal4 | M | 7037.20 | 0.71 |
| fru-M-400121 | 1011 | FruMARCM-M001189_seg001 | fru-Gal4 | M | 7312.86 | 0.69 |
| fru-M-400131 | 1119 | FruMARCM-M001317_seg002 | fru-Gal4 | M | 6879.66 | 0.69 |
| fru-M-400138 | 1196 | FruMARCM-M001376_seg001 | fru-Gal4 | M | 6974.15 | 0.70 |
| fru-M-400143 | 1240 | FruMARCM-M001421_seg001 | fru-Gal4 | M | 7217.47 | 0.61 |
| fru-M-300108 | 1491 | FruMARCM-M001045_seg001 | fru-Gal4 | M | 6997.25 | 0.71 |
| fru-M-700045 | 1664 | FruMARCM-M000813_seg002 | fru-Gal4 | M | 6972.70 | 0.69 |
| fru-M-400027 | 1815 | FruMARCM-M000338_seg001 | fru-Gal4 | M | 6694.34 | 0.64 |
| Trh-M-400062 | 1994 | TPHMARCM-1059M_seg001 | Trh-Gal4 | M | 7005.00 | 0.70 |
| Trh-M-400067 | 2835 | TPHMARCM-1062M_seg1 | Trh-Gal4 | M | 7013.68 | 0.56 |
| fru-F-300087 | 4499 | FruMARCM-F001602_seg001 | fru-Gal4 | F | 7269.27 | 0.69 |
| fru-F-400208 | 4777 | FruMARCM-F002094_seg001 | fru-Gal4 | F | 6796.70 | 0.65 |
| Cha-F-200177 | 5575 | ChaMARCM-F000872_seg002 | Cha-Gal4 | F | 7087.60 | 0.73 |
| VGlut-F-200600 | 5777 | DvGlutMARCM-F004481_seg001 | VGlut-Gal4 | F | 6930.28 | 0.68 |
| fru-F-300061 | 6184 | FruMARCM-F001117_seg001 | fru-Gal4 | F | 7091.18 | 0.73 |
| fru-F-400133 | 6189 | FruMARCM-F001120_seg002 | fru-Gal4 | F | 6699.43 | 0.70 |
| VGlut-F-500796 | 6501 | DvGlutMARCM-F004280_seg001 | VGlut-Gal4 | F | 7049.37 | 0.70 |
| VGlut-F-600759 | 6958 | DvGlutMARCM-F004110_seg001 | VGlut-Gal4 | F | 7031.44 | 0.65 |
| Cha-F-600084 | 7733 | ChaMARCM-F000541_seg002 | Cha-Gal4 | F | 7188.53 | 0.71 |
| Cha-F-400066 | 8386 | ChaMARCM-F000324_seg002 | Cha-Gal4 | F | 5136.52 | 0.41 |
| fru-F-700025 | 9777 | FruMARCM-F000296_seg001 | fru-Gal4 | F | 6864.06 | 0.65 |
| Trh-F-400087 | 12735 | TPHMARCM-957F_seg1 | Trh-Gal4 | F | 6527.72 | 0.65 |
| VGlut-F-400243 | 14579 | DvGlutMARCM-F845_seg1 | VGlut-Gal4 | F | 6411.95 | 0.65 |
| VGlut-F-500233 | 15349 | DvGlutMARCM-F1699_seg1 | VGlut-Gal4 | F | 7261.98 | 0.71 |