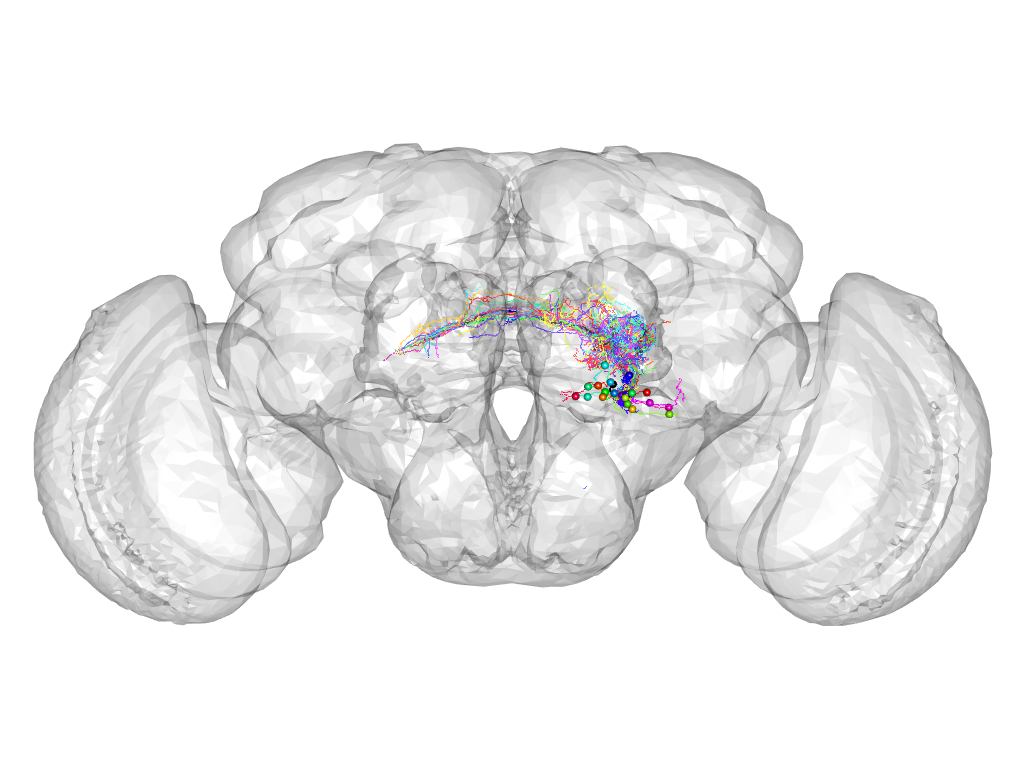
Cluster 420
Part of supercluster XII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-700517), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 653 | Trh-F-000043 | XIX | 0.67 |
| 555 | VGlut-F-900099 | XII | 0.66 |
| 662 | Tdc2-F-400026 | XIII | 0.64 |
| 997 | VGlut-F-400032 | XII | 0.60 |
| 400 | VGlut-F-500072 | XII | 0.60 |
| 774 | VGlut-F-500418 | XII | 0.57 |
| 806 | Trh-F-100085 | XII | 0.57 |
| 994 | VGlut-F-200045 | XIX | 0.55 |
| 144 | Trh-M-400113 | XII | 0.52 |
| 835 | VGlut-F-400504 | XII | 0.52 |
| 545 | VGlut-F-700278 | XII | 0.52 |
Cluster composition
This cluster has 25 neurons. The exemplar of this cluster (VGlut-F-700517) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-700517 | 6441 | DvGlutMARCM-F004228_seg001 | VGlut-Gal4 | F | 6548.81 | 1.00 |
| VGlut-F-600295 | 6029 | DvGlutMARCM-F002545_seg001 | VGlut-Gal4 | F | 4212.38 | 0.53 |
| VGlut-F-800262 | 6498 | DvGlutMARCM-F004277_seg001 | VGlut-Gal4 | F | 5166.41 | 0.77 |
| VGlut-F-800269 | 6515 | DvGlutMARCM-F004289_seg003 | VGlut-Gal4 | F | 4691.68 | 0.73 |
| VGlut-F-200555 | 7022 | DvGlutMARCM-F004159_seg001 | VGlut-Gal4 | F | 4393.76 | 0.62 |
| VGlut-F-700255 | 8469 | DvGlutMARCM-F003066_seg001 | VGlut-Gal4 | F | 4406.60 | 0.55 |
| VGlut-F-700292 | 8529 | DvGlutMARCM-F003107_seg001 | VGlut-Gal4 | F | 4743.20 | 0.74 |
| VGlut-F-400770 | 8688 | DvGlutMARCM-F003725_seg001 | VGlut-Gal4 | F | 4946.30 | 0.72 |
| VGlut-F-500620 | 9200 | DvGlutMARCM-F003267_seg001 | VGlut-Gal4 | F | 4863.25 | 0.69 |
| VGlut-F-500596 | 9377 | DvGlutMARCM-F003208_seg001 | VGlut-Gal4 | F | 4966.88 | 0.74 |
| VGlut-F-700212 | 10838 | DvGlutMARCM-F002839_seg001 | VGlut-Gal4 | F | 4750.28 | 0.72 |
| VGlut-F-600469 | 11092 | DvGlutMARCM-F002966_seg001 | VGlut-Gal4 | F | 4838.71 | 0.72 |
| VGlut-F-800117 | 11156 | DvGlutMARCM-F003009_seg001 | VGlut-Gal4 | F | 5011.68 | 0.74 |
| VGlut-F-600256 | 11860 | DvGlutMARCM-F002408_seg001 | VGlut-Gal4 | F | 4787.32 | 0.67 |
| VGlut-F-400536 | 11914 | DvGlutMARCM-F002449_seg001 | VGlut-Gal4 | F | 5133.00 | 0.77 |
| VGlut-F-600355 | 12188 | DvGlutMARCM-F002663_seg001 | VGlut-Gal4 | F | 5036.60 | 0.76 |
| VGlut-F-600034 | 14489 | DvGlutMARCM-F762_seg1 | VGlut-Gal4 | F | 5178.32 | 0.76 |
| VGlut-F-400220 | 14519 | DvGlutMARCM-F790_seg1 | VGlut-Gal4 | F | 5057.62 | 0.72 |
| VGlut-F-400222 | 14526 | DvGlutMARCM-F797-x2_seg1 | VGlut-Gal4 | F | 4320.53 | 0.58 |
| VGlut-F-500116 | 14601 | DvGlutMARCM-F863_seg1 | VGlut-Gal4 | F | 5000.47 | 0.73 |
| VGlut-F-500130 | 14615 | DvGlutMARCM-F874_seg1 | VGlut-Gal4 | F | 5186.81 | 0.76 |
| VGlut-F-400270 | 14653 | DvGlutMARCM-F907_seg1 | VGlut-Gal4 | F | 5026.68 | 0.71 |
| VGlut-F-400277 | 14673 | DvGlutMARCM-F934_seg1 | VGlut-Gal4 | F | 5063.31 | 0.73 |
| VGlut-F-000350 | 15040 | DvGlutMARCM-F1378_seg1 | VGlut-Gal4 | F | 5047.42 | 0.75 |
| VGlut-F-600070 | 15149 | DvGlutMARCM-F1483_seg2 | VGlut-Gal4 | F | 4780.63 | 0.69 |