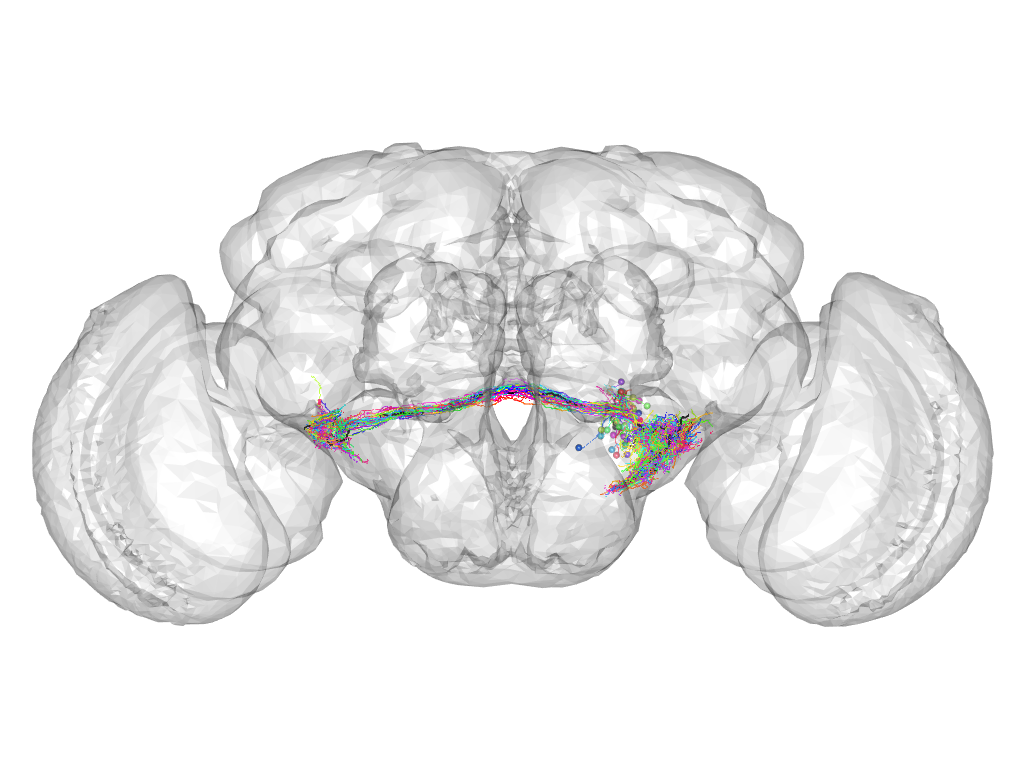
Cluster 309
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-700160), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 410 | fru-F-400147 | VIII | 0.70 |
| 27 | fru-M-300302 | VIII | 0.64 |
| 902 | VGlut-F-400300 | VIII | 0.62 |
| 320 | Cha-F-400233 | VIII | 0.56 |
| 459 | VGlut-F-500779 | VIII | 0.56 |
Cluster composition
This cluster has 37 neurons. The exemplar of this cluster (fru-F-700160) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-700160 | 4666 | FruMARCM-F001828_seg001 | fru-Gal4 | F | 13450.68 | 1.00 |
| fru-M-400218 | 253 | FruMARCM-M002481_seg001 | fru-Gal4 | M | 9917.34 | 0.75 |
| fru-M-300190 | 489 | FruMARCM-M001588_seg001 | fru-Gal4 | M | 9962.94 | 0.76 |
| fru-M-400175 | 724 | FruMARCM-M001934_seg001 | fru-Gal4 | M | 9522.35 | 0.70 |
| fru-M-400185 | 771 | FruMARCM-M001982_seg003 | fru-Gal4 | M | 10043.66 | 0.74 |
| fru-M-400190 | 816 | FruMARCM-M002022_seg001 | fru-Gal4 | M | 10086.65 | 0.73 |
| fru-M-400198 | 911 | FruMARCM-M002173_seg001 | fru-Gal4 | M | 9781.63 | 0.71 |
| fru-M-400135 | 1193 | FruMARCM-M001373_seg001 | fru-Gal4 | M | 9420.00 | 0.72 |
| fru-M-400145 | 1242 | FruMARCM-M001422_seg001 | fru-Gal4 | M | 9653.51 | 0.75 |
| npf-M-800000 | 2023 | npfMARCM-M000015_seg001 | npf-GAL4 | M | 10078.59 | 0.72 |
| fru-F-200159 | 3760 | FruMARCM-F002353_seg001 | fru-Gal4 | F | 9656.22 | 0.72 |
| fru-F-500277 | 3784 | FruMARCM-F002458_seg001 | fru-Gal4 | F | 9943.73 | 0.74 |
| fru-F-400234 | 3819 | FruMARCM-F002631_seg001 | fru-Gal4 | F | 9827.19 | 0.71 |
| fru-F-300086 | 4492 | FruMARCM-F001542_seg002 | fru-Gal4 | F | 10169.91 | 0.74 |
| fru-F-300097 | 4543 | FruMARCM-F001654_seg003 | fru-Gal4 | F | 9950.07 | 0.74 |
| fru-F-400197 | 4752 | FruMARCM-F002073_seg001 | fru-Gal4 | F | 9964.31 | 0.75 |
| Cha-F-400196 | 5580 | ChaMARCM-F000874_seg001 | Cha-Gal4 | F | 10114.27 | 0.74 |
| fru-F-700120 | 6227 | FruMARCM-F001167_seg001 | fru-Gal4 | F | 10027.64 | 0.75 |
| fru-F-400143 | 6281 | FruMARCM-F001311_seg001 | fru-Gal4 | F | 9586.71 | 0.73 |
| fru-F-800032 | 7287 | FruMARCM-F000917_seg002 | fru-Gal4 | F | 9371.81 | 0.73 |
| fru-F-700082 | 7294 | FruMARCM-F000923_seg002 | fru-Gal4 | F | 10057.21 | 0.75 |
| Cha-F-000044 | 8259 | ChaMARCM-F000244_seg001 | Cha-Gal4 | F | 9835.22 | 0.74 |
| npf-F-500002 | 9434 | npfMARCM-F000023_seg001 | npf-GAL4 | F | 9360.62 | 0.71 |
| npf-F-500004 | 9436 | npfMARCM-F000025_seg001 | npf-GAL4 | F | 10192.48 | 0.74 |
| npf-F-500005 | 9438 | npfMARCM-F000027_seg001 | npf-GAL4 | F | 9566.24 | 0.74 |
| npf-F-400000 | 9440 | npfMARCM-F000029_seg001 | npf-GAL4 | F | 9501.15 | 0.71 |
| npf-F-400001 | 9441 | npfMARCM-F000030_seg001 | npf-GAL4 | F | 9870.88 | 0.74 |
| npf-F-300007 | 9443 | npfMARCM-F000033_seg001 | npf-GAL4 | F | 9737.62 | 0.71 |
| npf-F-300008 | 9446 | npfMARCM-F000036_seg001 | npf-GAL4 | F | 9816.26 | 0.74 |
| npf-F-300010 | 9448 | npfMARCM-F000038_seg001 | npf-GAL4 | F | 9678.40 | 0.70 |
| npf-F-300011 | 9449 | npfMARCM-F000039_seg001 | npf-GAL4 | F | 9870.16 | 0.72 |
| fru-F-300027 | 9793 | FruMARCM-F000312_seg001 | fru-Gal4 | F | 9763.70 | 0.74 |
| npf-F-800004 | 10273 | npfMARCM-F000012_seg001 | npf-GAL4 | F | 9477.75 | 0.70 |
| npf-F-300005 | 10274 | npfMARCM-F000020_seg001 | npf-GAL4 | F | 9602.47 | 0.71 |
| npf-F-300004 | 11208 | npfMARCM-F000004_seg001 | npf-GAL4 | F | 9860.85 | 0.73 |
| npf-F-800000 | 11210 | npfMARCM-F000008_seg001 | npf-GAL4 | F | 10053.95 | 0.72 |
| fru-F-400013 | 11470 | FruMARCM-F000082_seg002 | fru-Gal4 | F | 9674.81 | 0.72 |