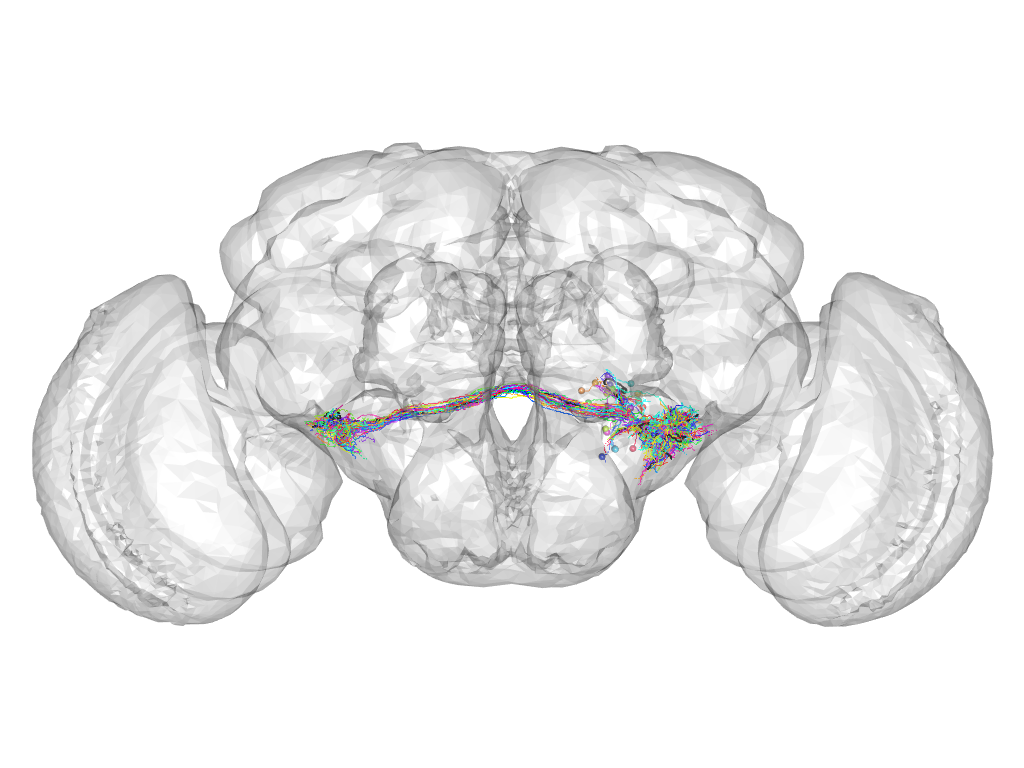
Cluster 27
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-300302), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 320 | Cha-F-400233 | VIII | 0.68 |
| 309 | fru-F-700160 | VIII | 0.67 |
| 410 | fru-F-400147 | VIII | 0.62 |
| 902 | VGlut-F-400300 | VIII | 0.60 |
| 459 | VGlut-F-500779 | VIII | 0.59 |
| 47 | fru-M-300198 | VIII | 0.58 |
Cluster composition
This cluster has 29 neurons. The exemplar of this cluster (fru-M-300302) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-300302 | 278 | FruMARCM-M002503_seg001 | fru-Gal4 | M | 13678.46 | 1.00 |
| fru-M-700144 | 37 | FruMARCM-M002233_seg001 | fru-Gal4 | M | 10280.04 | 0.75 |
| fru-M-400213 | 144 | FruMARCM-M002371_seg001 | fru-Gal4 | M | 9958.58 | 0.74 |
| fru-M-300266 | 179 | FruMARCM-M002395_seg002 | fru-Gal4 | M | 10275.61 | 0.75 |
| fru-M-400220 | 270 | FruMARCM-M002496_seg001 | fru-Gal4 | M | 10262.83 | 0.75 |
| fru-M-200217 | 501 | FruMARCM-M001596_seg001 | fru-Gal4 | M | 10145.21 | 0.74 |
| fru-M-400186 | 777 | FruMARCM-M001987_seg001 | fru-Gal4 | M | 9823.30 | 0.73 |
| fru-M-400193 | 819 | FruMARCM-M002024_seg001 | fru-Gal4 | M | 10098.68 | 0.74 |
| fru-M-300149 | 1152 | FruMARCM-M001344_seg001 | fru-Gal4 | M | 10142.31 | 0.74 |
| fru-M-400152 | 1298 | FruMARCM-M001492_seg001 | fru-Gal4 | M | 9731.51 | 0.72 |
| fru-M-300103 | 1455 | FruMARCM-M001021_seg001 | fru-Gal4 | M | 9916.47 | 0.73 |
| fru-M-200128 | 1540 | FruMARCM-M001074_seg001 | fru-Gal4 | M | 9747.89 | 0.74 |
| fru-M-300071 | 1572 | FruMARCM-M000644_seg001 | fru-Gal4 | M | 10119.65 | 0.74 |
| fru-M-400009 | 1790 | FruMARCM-M000318_seg002 | fru-Gal4 | M | 9832.31 | 0.73 |
| fru-F-300102 | 4616 | FruMARCM-F001742_seg001 | fru-Gal4 | F | 10331.48 | 0.72 |
| fru-F-600109 | 4652 | FruMARCM-F001816_seg001 | fru-Gal4 | F | 9623.48 | 0.69 |
| fru-F-700166 | 4674 | FruMARCM-F001832_seg002 | fru-Gal4 | F | 9913.27 | 0.72 |
| fru-F-400182 | 4718 | FruMARCM-F002044_seg001 | fru-Gal4 | F | 10229.88 | 0.75 |
| fru-F-000036 | 6162 | FruMARCM-F001103_seg001 | fru-Gal4 | F | 9404.36 | 0.66 |
| fru-F-700116 | 6208 | FruMARCM-F001133_seg001 | fru-Gal4 | F | 9839.13 | 0.71 |
| fru-F-700118 | 6221 | FruMARCM-F001157_seg001 | fru-Gal4 | F | 10467.67 | 0.76 |
| fru-F-200093 | 6314 | FruMARCM-F001434_seg001 | fru-Gal4 | F | 9685.77 | 0.72 |
| fru-F-700069 | 7276 | FruMARCM-F000912_seg001 | fru-Gal4 | F | 9900.18 | 0.72 |
| fru-F-500150 | 7290 | FruMARCM-F000920_seg001 | fru-Gal4 | F | 9661.71 | 0.73 |
| fru-F-400112 | 7317 | FruMARCM-F000956_seg001 | fru-Gal4 | F | 10003.56 | 0.74 |
| fru-F-400119 | 7339 | FruMARCM-F000976_seg001 | fru-Gal4 | F | 10212.92 | 0.74 |
| fru-F-500130 | 7806 | FruMARCM-F000704_seg001 | fru-Gal4 | F | 9670.89 | 0.72 |
| fru-F-400033 | 9738 | FruMARCM-F000254_seg001 | fru-Gal4 | F | 9689.60 | 0.72 |
| Trh-F-500183 | 12541 | TPHMARCM-1329F_seg1 | Trh-Gal4 | F | 9278.30 | 0.67 |