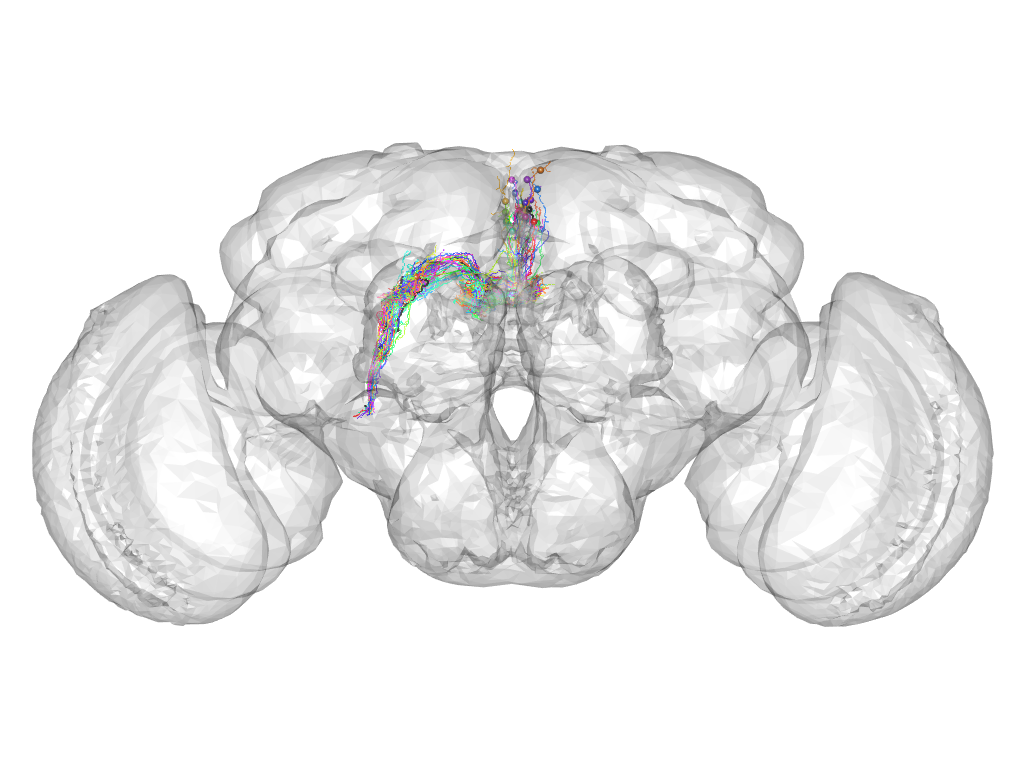
Cluster 231
Part of supercluster XIV
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Cha-F-300338), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 367 | Cha-F-000160 | XIV | 0.60 |
| 354 | Cha-F-100235 | XIV | 0.41 |
| 645 | fru-F-500100 | XIV | 0.38 |
| 658 | Tdc2-F-300026 | XIV | 0.34 |
| 416 | fru-F-600085 | XIV | 0.33 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (Cha-F-300338) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Cha-F-300338 | 3902 | ChaMARCM-F001563_seg001 | Cha-Gal4 | F | 28404.74 | 1.00 |
| Cha-F-700198 | 3932 | ChaMARCM-F001590_seg001 | Cha-Gal4 | F | 21879.44 | 0.77 |
| Gad1-F-800084 | 4088 | GadMARCM-F000676_seg001 | Gad1-Gal4 | F | 21938.31 | 0.77 |
| Cha-F-100256 | 4168 | ChaMARCM-F001279_seg001 | Cha-Gal4 | F | 20717.19 | 0.72 |
| Cha-F-300323 | 4461 | ChaMARCM-F001502_seg001 | Cha-Gal4 | F | 21270.96 | 0.72 |
| Cha-F-200194 | 4861 | ChaMARCM-F000983_seg001 | Cha-Gal4 | F | 22113.75 | 0.78 |
| Cha-F-300197 | 5302 | ChaMARCM-F000686_seg001 | Cha-Gal4 | F | 20180.43 | 0.72 |
| Cha-F-000125 | 5348 | ChaMARCM-F000720_seg001 | Cha-Gal4 | F | 20029.36 | 0.70 |
| Cha-F-400170 | 5394 | ChaMARCM-F000753_seg001 | Cha-Gal4 | F | 20658.98 | 0.72 |
| Cha-F-500150 | 5445 | ChaMARCM-F000785_seg001 | Cha-Gal4 | F | 21573.41 | 0.78 |
| Cha-F-700146 | 5457 | ChaMARCM-F000794_seg001 | Cha-Gal4 | F | 19720.12 | 0.72 |
| Cha-F-800032 | 5462 | ChaMARCM-F000798_seg001 | Cha-Gal4 | F | 21183.53 | 0.74 |
| Cha-F-200175 | 5573 | ChaMARCM-F000871_seg001 | Cha-Gal4 | F | 20835.04 | 0.72 |
| Cha-F-400129 | 7125 | ChaMARCM-F000594_seg001 | Cha-Gal4 | F | 19522.94 | 0.64 |
| Cha-F-300160 | 7139 | ChaMARCM-F000606_seg001 | Cha-Gal4 | F | 20893.71 | 0.74 |
| Cha-F-600092 | 7180 | ChaMARCM-F000632_seg001 | Cha-Gal4 | F | 20977.69 | 0.74 |
| Cha-F-400098 | 7564 | ChaMARCM-F000429_seg001 | Cha-Gal4 | F | 19852.12 | 0.72 |
| Cha-F-000031 | 8073 | ChaMARCM-F000140_seg001 | Cha-Gal4 | F | 20999.63 | 0.73 |
| Gad1-F-600050 | 9465 | GadMARCM-F000366_seg001 | Gad1-Gal4 | F | 21040.55 | 0.74 |
| Gad1-F-200106 | 9534 | GadMARCM-F000417_seg001 | Gad1-Gal4 | F | 21012.90 | 0.74 |
| Gad1-F-400077 | 9548 | GadMARCM-F000428_seg001 | Gad1-Gal4 | F | 22152.37 | 0.78 |
| Gad1-F-300111 | 9578 | GadMARCM-F000449_seg001 | Gad1-Gal4 | F | 21461.18 | 0.76 |
| Gad1-F-000054 | 9655 | GadMARCM-F000506_seg001 | Gad1-Gal4 | F | 20440.09 | 0.72 |
| Gad1-F-800059 | 9692 | GadMARCM-F000541_seg001 | Gad1-Gal4 | F | 19951.79 | 0.71 |
| Cha-F-400029 | 10131 | ChaMARCM-F000077_seg001 | Cha-Gal4 | F | 21807.39 | 0.78 |
| Cha-F-200013 | 10147 | ChaMARCM-F000086_seg001 | Cha-Gal4 | F | 21813.57 | 0.77 |
| Gad1-F-700024 | 11291 | GadMARCM-F000263_seg001 | Gad1-Gal4 | F | 19359.01 | 0.73 |
| Gad1-F-500065 | 11353 | GadMARCM-F000313_seg001 | Gad1-Gal4 | F | 21932.52 | 0.77 |