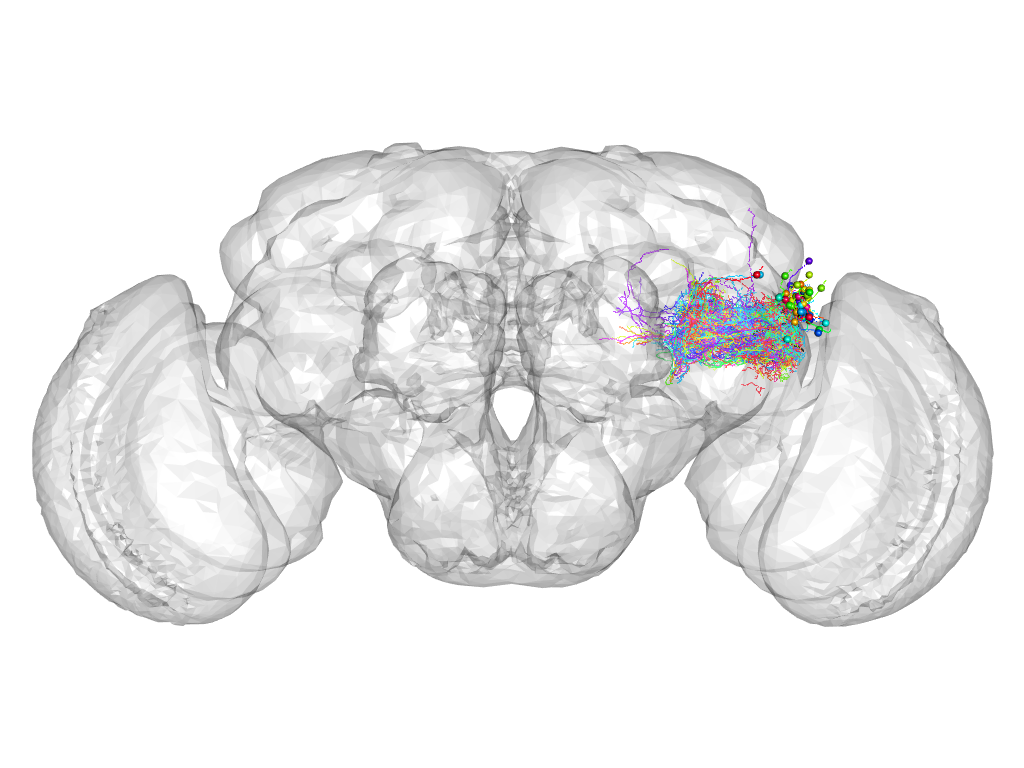
Cluster 216
Part of supercluster X
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-F-800061), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 750 | VGlut-F-400641 | X | 0.64 |
| 390 | VGlut-F-600386 | X | 0.63 |
| 321 | Cha-F-600151 | X | 0.61 |
| 92 | fru-M-100103 | X | 0.59 |
| 674 | Gad1-F-600005 | X | 0.58 |
| 939 | VGlut-F-400341 | X | 0.57 |
Cluster composition
This cluster has 68 neurons. The exemplar of this cluster (fru-F-800061) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-F-800061 | 3721 | FruMARCM-F002220_seg001 | fru-Gal4 | F | 17733.03 | 1.00 |
| fru-M-200262 | 49 | FruMARCM-M002244_seg001 | fru-Gal4 | M | 12873.76 | 0.71 |
| fru-M-600102 | 88 | FruMARCM-M002288_seg001 | fru-Gal4 | M | 8521.31 | 0.51 |
| fru-M-500157 | 546 | FruMARCM-M001683_seg001 | fru-Gal4 | M | 12892.15 | 0.72 |
| fru-M-500165 | 572 | FruMARCM-M001762_seg001 | fru-Gal4 | M | 12547.37 | 0.66 |
| fru-M-700123 | 791 | FruMARCM-M002005_seg001 | fru-Gal4 | M | 12391.67 | 0.69 |
| fru-M-500108 | 1020 | FruMARCM-M001203_seg002 | fru-Gal4 | M | 11724.27 | 0.69 |
| fru-M-500109 | 1021 | FruMARCM-M001204_seg001 | fru-Gal4 | M | 12147.42 | 0.65 |
| fru-M-500121 | 1055 | FruMARCM-M001234_seg001 | fru-Gal4 | M | 11675.78 | 0.68 |
| fru-M-200159 | 1073 | FruMARCM-M001249_seg002 | fru-Gal4 | M | 11296.42 | 0.65 |
| fru-M-000044 | 1112 | FruMARCM-M001293_seg001 | fru-Gal4 | M | 10300.91 | 0.59 |
| fru-M-000060 | 1166 | FruMARCM-M001352_seg001 | fru-Gal4 | M | 11642.61 | 0.70 |
| fru-M-600064 | 1267 | FruMARCM-M001457_seg001 | fru-Gal4 | M | 11966.60 | 0.68 |
| fru-M-600037 | 1411 | FruMARCM-M000907_seg003 | fru-Gal4 | M | 11833.93 | 0.68 |
| fru-M-400067 | 1418 | FruMARCM-M000932_seg001 | fru-Gal4 | M | 12629.01 | 0.64 |
| fru-M-200129 | 1541 | FruMARCM-M001075_seg001 | fru-Gal4 | M | 11270.17 | 0.60 |
| fru-M-500097 | 1556 | FruMARCM-M001088_seg001 | fru-Gal4 | M | 12054.78 | 0.67 |
| fru-M-300074 | 1609 | FruMARCM-M000725_seg001 | fru-Gal4 | M | 11506.38 | 0.69 |
| fru-M-700047 | 1666 | FruMARCM-M000814_seg001 | fru-Gal4 | M | 12106.92 | 0.69 |
| fru-M-500001 | 1757 | FruMARCM-M000240_seg001 | fru-Gal4 | M | 12483.88 | 0.70 |
| fru-M-500004 | 1829 | FruMARCM-M000347_seg001 | fru-Gal4 | M | 12676.10 | 0.70 |
| fru-M-500013 | 1845 | FruMARCM-M000356_seg001 | fru-Gal4 | M | 11913.20 | 0.69 |
| fru-M-500015 | 1848 | FruMARCM-M000358_seg002 | fru-Gal4 | M | 11708.45 | 0.69 |
| fru-M-100048 | 1918 | FruMARCM-M000475_seg001 | fru-Gal4 | M | 11825.87 | 0.70 |
| fru-M-200023 | 2349 | FruMARCM-M000042_seg002 | fru-Gal4 | M | 11397.31 | 0.63 |
| fru-F-600124 | 3719 | FruMARCM-F002217_seg001 | fru-Gal4 | F | 12533.27 | 0.71 |
| fru-F-800067 | 3800 | FruMARCM-F002512_seg002 | fru-Gal4 | F | 8765.69 | 0.53 |
| Cha-F-600164 | 4172 | ChaMARCM-F001280_seg003 | Cha-Gal4 | F | 11477.82 | 0.66 |
| fru-F-600092 | 4507 | FruMARCM-F001615_seg001 | fru-Gal4 | F | 10855.92 | 0.62 |
| fru-F-500195 | 4513 | FruMARCM-F001623_seg001 | fru-Gal4 | F | 12281.49 | 0.69 |
| fru-F-800047 | 4517 | FruMARCM-F001627_seg001 | fru-Gal4 | F | 7367.17 | 0.56 |
| fru-F-600095 | 4561 | FruMARCM-F001670_seg001 | fru-Gal4 | F | 12161.09 | 0.69 |
| fru-F-200119 | 4613 | FruMARCM-F001734_seg002 | fru-Gal4 | F | 11054.72 | 0.63 |
| fru-F-500236 | 4711 | FruMARCM-F002039_seg001 | fru-Gal4 | F | 12716.51 | 0.71 |
| fru-F-500242 | 4733 | FruMARCM-F002058_seg002 | fru-Gal4 | F | 12451.15 | 0.70 |
| fru-F-700176 | 4821 | FruMARCM-F002186_seg001 | fru-Gal4 | F | 13181.72 | 0.74 |
| Cha-F-400191 | 5558 | ChaMARCM-F000858_seg001 | Cha-Gal4 | F | 11794.75 | 0.61 |
| Cha-F-700164 | 5602 | ChaMARCM-F000890_seg001 | Cha-Gal4 | F | 11930.07 | 0.57 |
| Cha-F-400202 | 5617 | ChaMARCM-F000898_seg001 | Cha-Gal4 | F | 12683.01 | 0.71 |
| fru-F-400137 | 6199 | FruMARCM-F001127_seg001 | fru-Gal4 | F | 12160.81 | 0.68 |
| fru-F-500165 | 6232 | FruMARCM-F001193_seg001 | fru-Gal4 | F | 12012.56 | 0.70 |
| fru-F-600067 | 6248 | FruMARCM-F001259_seg001 | fru-Gal4 | F | 12195.57 | 0.70 |
| fru-F-600070 | 6285 | FruMARCM-F001335_seg001 | fru-Gal4 | F | 12090.85 | 0.69 |
| fru-F-500182 | 6320 | FruMARCM-F001442_seg001 | fru-Gal4 | F | 11519.23 | 0.63 |
| fru-F-600082 | 6363 | FruMARCM-F001504_seg002 | fru-Gal4 | F | 11668.25 | 0.70 |
| fru-F-700155 | 6381 | FruMARCM-F001518_seg001 | fru-Gal4 | F | 9293.47 | 0.52 |
| VGlut-F-500835 | 6625 | DvGlutMARCM-F004377_seg002 | VGlut-Gal4 | F | 11991.72 | 0.71 |
| Cha-F-400123 | 7117 | ChaMARCM-F000590_seg001 | Cha-Gal4 | F | 10592.83 | 0.60 |
| fru-F-800036 | 7301 | FruMARCM-F000930_seg001 | fru-Gal4 | F | 10374.71 | 0.56 |
| fru-F-700094 | 7329 | FruMARCM-F000966_seg001 | fru-Gal4 | F | 12184.75 | 0.71 |
| Cha-F-000066 | 7462 | ChaMARCM-F000341_seg001 | Cha-Gal4 | F | 11066.48 | 0.60 |
| Cha-F-200074 | 7593 | ChaMARCM-F000445_seg001 | Cha-Gal4 | F | 11323.63 | 0.60 |
| Cha-F-000091 | 7703 | ChaMARCM-F000521_seg001 | Cha-Gal4 | F | 10122.15 | 0.56 |
| Cha-F-000042 | 8103 | ChaMARCM-F000158_seg001 | Cha-Gal4 | F | 8644.22 | 0.39 |
| Cha-F-900009 | 8333 | ChaMARCM-F000287_seg003 | Cha-Gal4 | F | 10922.90 | 0.66 |
| fru-F-400038 | 9743 | FruMARCM-F000259_seg001 | fru-Gal4 | F | 11772.18 | 0.65 |
| fru-F-400040 | 9745 | FruMARCM-F000259_seg003 | fru-Gal4 | F | 11519.26 | 0.61 |
| fru-F-700029 | 9781 | FruMARCM-F000298_seg001 | fru-Gal4 | F | 12162.97 | 0.72 |
| fru-F-700042 | 9902 | FruMARCM-F000491_seg001 | fru-Gal4 | F | 11715.49 | 0.70 |
| fru-F-500024 | 9905 | FruMARCM-F000492_seg002 | fru-Gal4 | F | 10327.83 | 0.56 |
| fru-F-600027 | 9946 | FruMARCM-F000531_seg001 | fru-Gal4 | F | 12988.76 | 0.73 |
| fru-F-500038 | 9948 | FruMARCM-F000542_seg001 | fru-Gal4 | F | 12196.95 | 0.71 |
| fru-F-500045 | 9961 | FruMARCM-F000552_seg001 | fru-Gal4 | F | 12449.53 | 0.71 |
| fru-F-500046 | 9962 | FruMARCM-F000553_seg001 | fru-Gal4 | F | 12259.65 | 0.70 |
| fru-F-600040 | 10008 | FruMARCM-F000588_seg001 | fru-Gal4 | F | 12400.65 | 0.72 |
| fru-F-600003 | 11491 | FruMARCM-F000156_seg001 | fru-Gal4 | F | 11905.73 | 0.66 |
| fru-F-400031 | 11514 | FruMARCM-F000175_seg001 | fru-Gal4 | F | 11763.22 | 0.69 |
| Trh-F-500218 | 12609 | TPHMARCM-F001404_seg001 | Trh-Gal4 | F | 11206.85 | 0.56 |