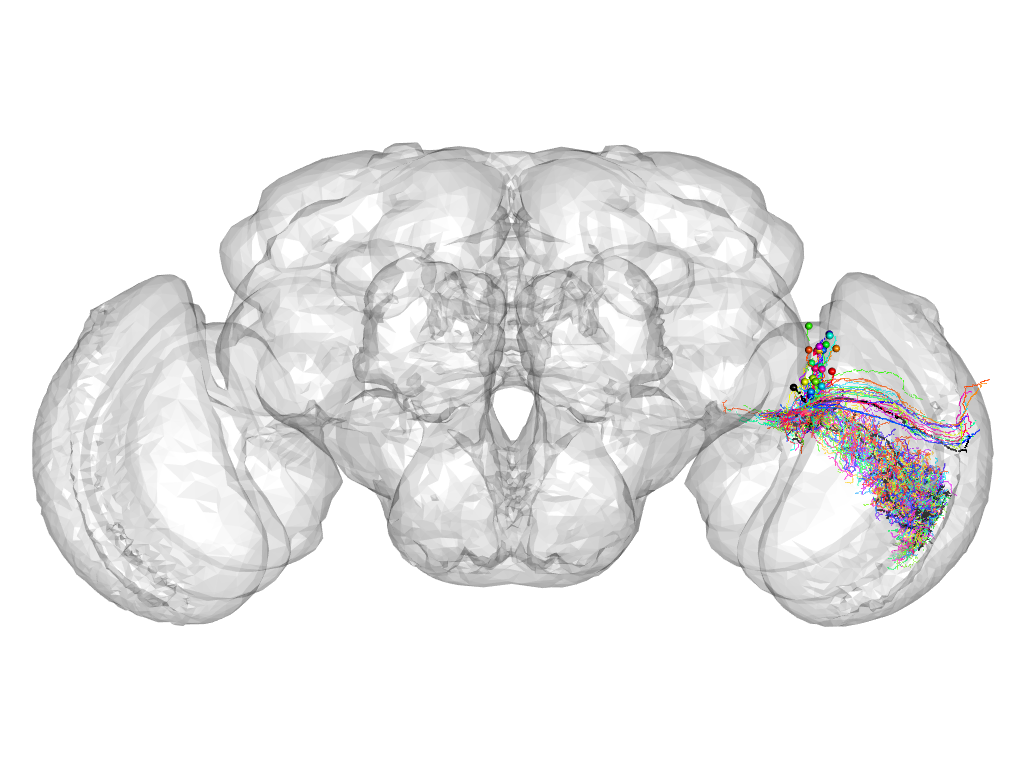
Cluster 208
Part of supercluster III
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (Trh-M-500030), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 661 | Tdc2-F-000027 | VII | 0.68 |
| 64 | fru-M-300237 | III | 0.66 |
| 76 | fru-M-300240 | III | 0.63 |
| 173 | Trh-M-000051 | III | 0.62 |
| 952 | VGlut-F-600052 | III | 0.62 |
| 1006 | VGlut-F-500043 | III | 0.56 |
| 759 | VGlut-F-600257 | III | 0.55 |
| 209 | Trh-M-600023 | III | 0.54 |
| 468 | VGlut-F-600670 | III | 0.51 |
Cluster composition
This cluster has 24 neurons. The exemplar of this cluster (Trh-M-500030) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| Trh-M-500030 | 3597 | TPHMARCM-540M_seg1 | Trh-Gal4 | M | 23336.53 | 1.00 |
| fru-M-800071 | 612 | FruMARCM-M001801_seg001 | fru-Gal4 | M | 14136.40 | 0.63 |
| fru-M-000150 | 843 | FruMARCM-M002097_seg001 | fru-Gal4 | M | 14996.17 | 0.61 |
| Trh-M-900036 | 2046 | TPHMARCM-M001553_seg001 | Trh-Gal4 | M | 15135.79 | 0.63 |
| Trh-M-900050 | 2062 | TPHMARCM-M001567_seg001 | Trh-Gal4 | M | 14814.20 | 0.64 |
| Trh-M-600093 | 2190 | TPHMARCM-M001680_seg001 | Trh-Gal4 | M | 15303.13 | 0.68 |
| Trh-M-000062 | 2201 | TPHMARCM-M001691_seg001 | Trh-Gal4 | M | 15025.31 | 0.66 |
| Trh-M-700087 | 2244 | TPHMARCM-M001731_seg001 | Trh-Gal4 | M | 13637.71 | 0.62 |
| Trh-M-000120 | 2306 | TPHMARCM-M001788_seg001 | Trh-Gal4 | M | 14198.85 | 0.61 |
| fru-M-900008 | 2373 | FruMARCM-M000104_seg002 | fru-Gal4 | M | 14172.28 | 0.63 |
| Trh-M-600049 | 3009 | TPHMARCM-961M_seg1 | Trh-Gal4 | M | 15943.85 | 0.68 |
| Trh-M-200002 | 3419 | TPHMARCM-106M_seg1 | Trh-Gal4 | M | 14352.31 | 0.64 |
| Trh-M-400017 | 3508 | TPHMARCM-369M_seg4 | Trh-Gal4 | M | 15467.30 | 0.68 |
| Trh-M-600007 | 3581 | TPHMARCM-519M_seg1 | Trh-Gal4 | M | 15605.82 | 0.67 |
| Trh-M-600019 | 3609 | TPHMARCM-561M-01_seg1 | Trh-Gal4 | M | 12529.47 | 0.57 |
| fru-F-700171 | 4740 | FruMARCM-F002064_seg001 | fru-Gal4 | F | 15808.03 | 0.68 |
| fru-F-800060 | 4805 | FruMARCM-F002144_seg001 | fru-Gal4 | F | 15178.83 | 0.67 |
| fru-F-700177 | 4822 | FruMARCM-F002187_seg001 | fru-Gal4 | F | 15804.39 | 0.67 |
| fru-F-500004 | 9752 | FruMARCM-F000266_seg001 | fru-Gal4 | F | 14685.52 | 0.65 |
| fru-F-300012 | 11525 | FruMARCM-F000184_seg001 | fru-Gal4 | F | 12571.96 | 0.56 |
| Trh-F-100062 | 12732 | TPHMARCM-953F_seg1 | Trh-Gal4 | F | 15948.68 | 0.69 |
| Trh-F-300007 | 13886 | TPHMARCM-151F_seg1 | Trh-Gal4 | F | 15491.69 | 0.64 |
| Trh-F-300014 | 13902 | TPHMARCM-166F_seg1 | Trh-Gal4 | F | 16058.58 | 0.69 |
| Trh-F-700040 | 14294 | TPHMARCM-835F_seg1 | Trh-Gal4 | F | 14724.98 | 0.64 |