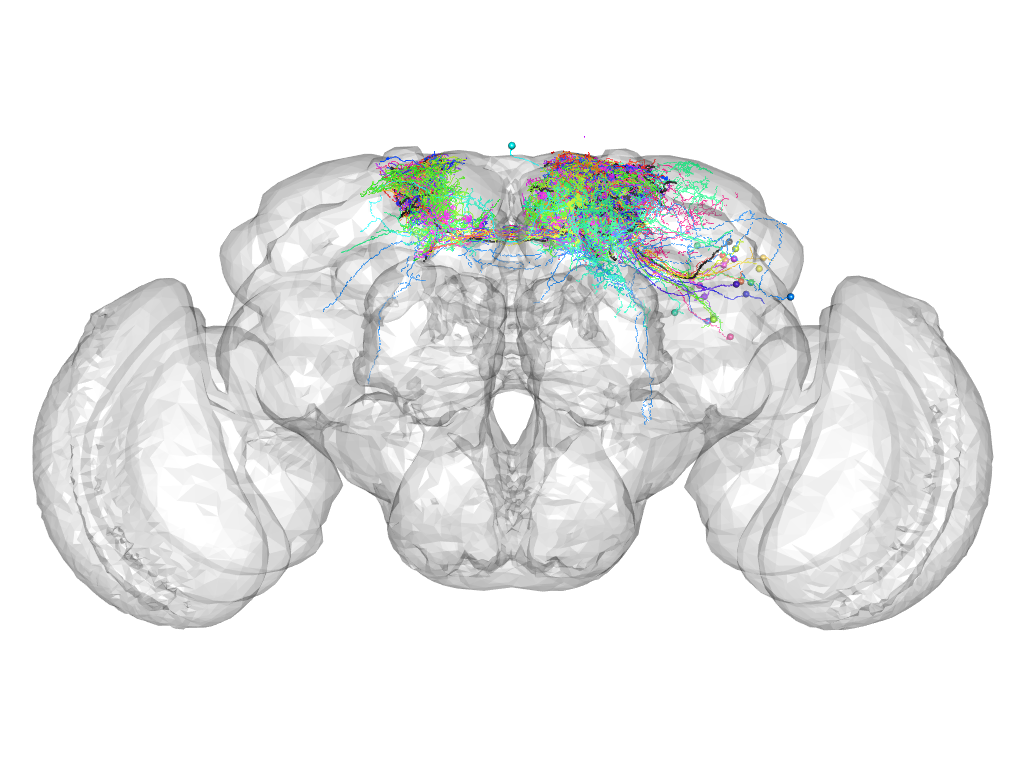
Cluster 193
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (TH-M-100039), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 196 | TH-M-300047 | XIX | 0.61 |
| 881 | Trh-F-200023 | XIX | 0.60 |
| 1003 | VGlut-F-000024 | XIX | 0.58 |
| 853 | TH-F-200037 | XIX | 0.57 |
| 143 | Trh-M-400111 | XIX | 0.55 |
| 83 | TH-M-300024 | XIX | 0.55 |
| 992 | VGlut-F-200024 | XIX | 0.55 |
| 190 | TH-M-200014 | XIX | 0.51 |
Cluster composition
This cluster has 25 neurons. The exemplar of this cluster (TH-M-100039) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| TH-M-100039 | 3184 | THMARCM-278M_seg1 | TH-Gal4 | M | 101717.21 | 1.00 |
| TH-M-300000 | 3077 | THMARCM-039M_seg1 | TH-Gal4 | M | 76930.79 | 0.75 |
| TH-M-200058 | 3239 | THMARCM-395M_seg1 | TH-Gal4 | M | 75631.08 | 0.75 |
| TH-M-200071 | 3260 | THMARCM-445M_seg1 | TH-Gal4 | M | 76037.35 | 0.74 |
| TH-M-300053 | 3293 | THMARCM-494M_seg1 | TH-Gal4 | M | 75799.62 | 0.76 |
| TH-M-300054 | 3308 | THMARCM-540M_seg1 | TH-Gal4 | M | 75194.09 | 0.76 |
| TH-M-300056 | 3311 | THMARCM-546M_seg1 | TH-Gal4 | M | 78671.11 | 0.76 |
| TH-M-000038 | 3335 | THMARCM-610M_seg | TH-Gal4 | M | 76420.01 | 0.70 |
| TH-M-000039 | 3338 | THMARCM-621M_seg | TH-Gal4 | M | 74925.75 | 0.70 |
| TH-M-000049 | 3347 | THMARCM-646M_seg | TH-Gal4 | M | 78663.14 | 0.73 |
| TH-M-300063 | 3363 | THMARCM-773M_seg1 | TH-Gal4 | M | 77592.49 | 0.76 |
| Cha-F-300267 | 4849 | ChaMARCM-F000973_seg001 | Cha-Gal4 | F | 63085.96 | 0.56 |
| Cha-F-100135 | 4880 | ChaMARCM-F000997_seg001 | Cha-Gal4 | F | 60527.04 | 0.49 |
| VGlut-F-700580 | 5843 | DvGlutMARCM-F004529_seg001 | VGlut-Gal4 | F | 45497.05 | 0.50 |
| VGlut-F-200383 | 6116 | DvGlutMARCM-F2194_seg2 | VGlut-Gal4 | F | 47069.16 | 0.52 |
| 5-HT1B-F-300000 | 11604 | 5HT1bMARCM-F000001_seg001 | 5-HT1B-Gal4 | F | 58375.12 | 0.40 |
| TH-F-300020 | 13403 | THMARCM-195F_seg1 | TH-Gal4 | F | 78407.15 | 0.74 |
| TH-F-200026 | 13459 | THMARCM-297F_seg1 | TH-Gal4 | F | 78930.55 | 0.78 |
| TH-F-200030 | 13466 | THMARCM-302F_seg1 | TH-Gal4 | F | 74330.58 | 0.72 |
| TH-F-100052 | 13562 | THMARCM-462F_seg2 | TH-Gal4 | F | 76659.51 | 0.76 |
| TH-F-300073 | 13580 | THMARCM-507F_seg1 | TH-Gal4 | F | 79196.92 | 0.78 |
| TH-F-000053 | 13662 | THMARCM-676F-X2_seg1 | TH-Gal4 | F | 73406.60 | 0.70 |
| TH-F-200116 | 13697 | THMARCM-719F_seg | TH-Gal4 | F | 75904.50 | 0.73 |
| Trh-F-000017 | 13915 | TPHMARCM-198F_seg1 | Trh-Gal4 | F | 63892.76 | 0.55 |
| VGlut-F-200020 | 15626 | DvGlutMARCM-F043-X3_seg1 | VGlut-Gal4 | F | 47569.81 | 0.53 |