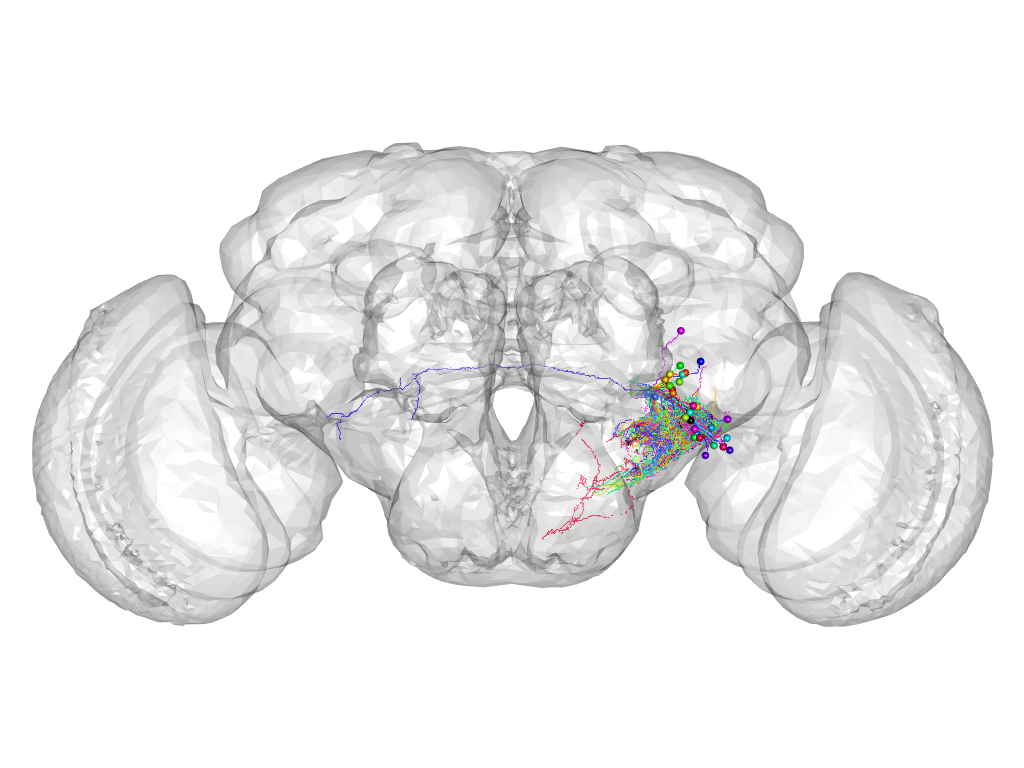
Cluster 159
Part of supercluster VIII
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (fru-M-700004), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 734 | fru-F-800001 | VIII | 0.66 |
| 988 | VGlut-F-200005 | X | 0.63 |
| 476 | fru-F-700099 | VIII | 0.59 |
| 406 | fru-F-000053 | VII | 0.59 |
| 652 | Tdc2-F-200010 | VIII | 0.59 |
| 12 | fru-M-500228 | VIII | 0.58 |
| 187 | TH-M-400003 | VIII | 0.57 |
| 27 | fru-M-300302 | VIII | 0.54 |
| 855 | TH-F-200057 | VIII | 0.53 |
| 962 | VGlut-F-600088 | X | 0.50 |
Cluster composition
This cluster has 28 neurons. The exemplar of this cluster (fru-M-700004) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| fru-M-700004 | 2428 | FruMARCM-M000148_seg001 | fru-Gal4 | M | 10728.65 | 1.00 |
| fru-M-100238 | 406 | FruMARCM-M002646_seg001 | fru-Gal4 | M | 7859.00 | 0.73 |
| fru-M-000114 | 603 | FruMARCM-M001793_seg002 | fru-Gal4 | M | 7565.67 | 0.72 |
| fru-M-400167 | 616 | FruMARCM-M001804_seg001 | fru-Gal4 | M | 7686.42 | 0.70 |
| fru-M-600084 | 679 | FruMARCM-M001884_seg001 | fru-Gal4 | M | 7488.59 | 0.71 |
| fru-M-000153 | 847 | FruMARCM-M002099_seg002 | fru-Gal4 | M | 7744.29 | 0.72 |
| fru-M-100120 | 1154 | FruMARCM-M001346_seg001 | fru-Gal4 | M | 7129.15 | 0.65 |
| fru-M-100055 | 1586 | FruMARCM-M000654_seg001 | fru-Gal4 | M | 7849.15 | 0.74 |
| fru-M-900024 | 1662 | FruMARCM-M000811_seg001 | fru-Gal4 | M | 7694.80 | 0.72 |
| fru-M-100044 | 1883 | FruMARCM-M000421_seg003 | fru-Gal4 | M | 7676.86 | 0.71 |
| fru-M-600024 | 1954 | FruMARCM-M000598_seg001 | fru-Gal4 | M | 7348.33 | 0.67 |
| fru-M-300014 | 2396 | FruMARCM-M000124_seg001 | fru-Gal4 | M | 7729.14 | 0.71 |
| Cha-F-200295 | 4207 | ChaMARCM-F001298_seg004 | Cha-Gal4 | F | 7774.44 | 0.68 |
| fru-F-200106 | 4472 | FruMARCM-F001663_seg001 | fru-Gal4 | F | 7564.34 | 0.70 |
| fru-F-000084 | 4558 | FruMARCM-F001668_seg001 | fru-Gal4 | F | 7572.98 | 0.72 |
| fru-F-500212 | 4595 | FruMARCM-F001712_seg001 | fru-Gal4 | F | 7502.89 | 0.70 |
| fru-F-000099 | 4641 | FruMARCM-F001808_seg001 | fru-Gal4 | F | 8006.74 | 0.74 |
| fru-F-900023 | 4680 | FruMARCM-F001854_seg001 | fru-Gal4 | F | 7731.06 | 0.72 |
| Cha-F-600068 | 7628 | ChaMARCM-F000467_seg001 | Cha-Gal4 | F | 7348.30 | 0.69 |
| fru-F-000029 | 7789 | FruMARCM-F000686_seg001 | fru-Gal4 | F | 7229.12 | 0.67 |
| VGlut-F-500571 | 8559 | DvGlutMARCM-F003125_seg002 | VGlut-Gal4 | F | 7573.26 | 0.47 |
| VGlut-F-700330 | 9294 | DvGlutMARCM-F003341_seg002 | VGlut-Gal4 | F | 7829.16 | 0.73 |
| fru-F-900006 | 9796 | FruMARCM-F000337_seg001 | fru-Gal4 | F | 7654.79 | 0.72 |
| fru-F-700043 | 9903 | FruMARCM-F000491_seg002 | fru-Gal4 | F | 7529.25 | 0.70 |
| fru-F-700011 | 11513 | FruMARCM-F000174_seg001 | fru-Gal4 | F | 7936.38 | 0.72 |
| VGlut-F-600102 | 12913 | DvGlutMARCM-F1953_seg1 | VGlut-Gal4 | F | 7741.34 | 0.72 |
| VGlut-F-600063 | 15117 | DvGlutMARCM-F1451_seg1 | VGlut-Gal4 | F | 7413.34 | 0.71 |
| VGlut-F-000138 | 16155 | DvGlutMARCM-F537_seg1 | VGlut-Gal4 | F | 8318.68 | 0.62 |