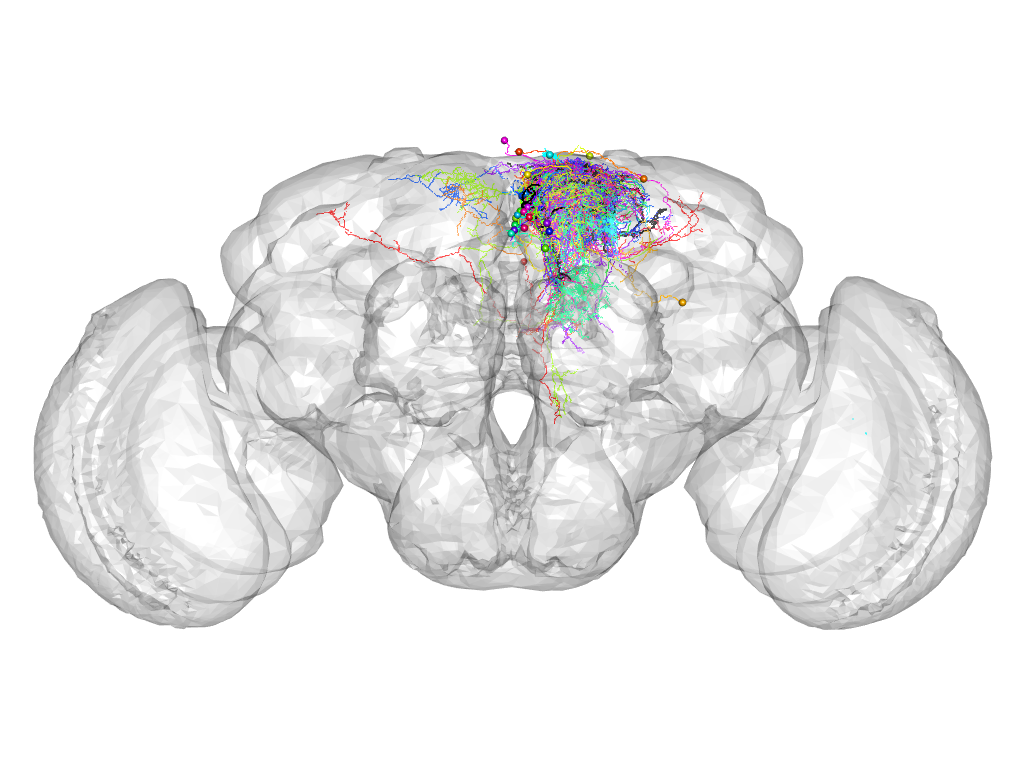
Cluster 1032
Part of supercluster XIX
About
Scores are NBLAST (version 2) similarity scores between each neuron and the exemplar (VGlut-F-000077), normalised to the self-score of the exemplar. The higher the better! Scores > 0 indicate a match of some sort. Excellent matches will have scores > 0.5.
Do you recognise any of the neurons in this cluster? Email us!
Credits
Raw data provided by flycircuit.tw as described in Three-Dimensional Reconstruction of Brain-wide Wiring Networks in Drosophila at Single-Cell Resolution by Ann-Shyn Chiang et al.
Image processing carried out with unu, CMTK, and Fiji.
Further processing carried out in R with nat. Clustering used package apcluster. 3D visualisation written from rgl by writeWebGL.
Similar clusters
The most similar clusters to this one are:
| Cluster | Exemplar | Supercluster | Exemplar normalised score |
|---|---|---|---|
| 36 | fru-M-200340 | XIX | 0.70 |
| 197 | TH-M-200095 | XIX | 0.63 |
| 862 | TH-F-200095 | XIX | 0.62 |
| 941 | VGlut-F-200251 | XIX | 0.62 |
| 204 | Trh-M-100025 | XIV | 0.62 |
| 1003 | VGlut-F-000024 | XIX | 0.58 |
| 198 | TH-M-000032 | XIX | 0.57 |
| 203 | Trh-M-000010 | XVIII | 0.55 |
| 859 | TH-F-200089 | XIV | 0.55 |
| 136 | TH-M-000013 | XVII | 0.53 |
| 186 | TH-M-600001 | XIX | 0.51 |
Cluster composition
This cluster has 27 neurons. The exemplar of this cluster (VGlut-F-000077) is drawn in black.
| Neuron | idid | Gene name | Driver | Sex | Raw score | Normalised score |
|---|---|---|---|---|---|---|
| VGlut-F-000077 | 16014 | DvGlutMARCM-F402_seg1 | VGlut-Gal4 | F | 103767.27 | 1.00 |
| fru-M-100052 | 1583 | FruMARCM-M000652_seg002 | fru-Gal4 | M | 55322.04 | 0.39 |
| fru-M-600021 | 1853 | FruMARCM-M000370_seg001 | fru-Gal4 | M | 55624.82 | 0.57 |
| fru-M-100002 | 2352 | FruMARCM-M000045_seg001 | fru-Gal4 | M | 56306.78 | 0.58 |
| fru-F-400164 | 4516 | FruMARCM-F001625_seg002 | fru-Gal4 | F | 35049.15 | 0.41 |
| fru-F-000097 | 4630 | FruMARCM-F001759_seg002 | fru-Gal4 | F | 57585.27 | 0.64 |
| fru-F-500250 | 4779 | FruMARCM-F002096_seg001 | fru-Gal4 | F | 58550.18 | 0.64 |
| Cha-F-300251 | 5624 | ChaMARCM-F000906_seg001 | Cha-Gal4 | F | 63195.84 | 0.49 |
| VGlut-F-100393 | 5778 | DvGlutMARCM-F004482_seg001 | VGlut-Gal4 | F | 76870.10 | 0.76 |
| Trh-F-300130 | 5985 | TPHMARCM-1004F_seg2 | Trh-Gal4 | F | 52236.04 | 0.62 |
| fru-F-300072 | 6213 | FruMARCM-F001137_seg001 | fru-Gal4 | F | 60180.60 | 0.63 |
| VGlut-F-400854 | 6884 | DvGlutMARCM-F004042_seg001 | VGlut-Gal4 | F | 73702.05 | 0.73 |
| VGlut-F-500768 | 6976 | DvGlutMARCM-F004122_seg001 | VGlut-Gal4 | F | 65326.57 | 0.60 |
| VGlut-F-200558 | 7060 | DvGlutMARCM-F004189_seg001 | VGlut-Gal4 | F | 72051.77 | 0.72 |
| Cha-F-500051 | 8143 | ChaMARCM-F000179_seg001 | Cha-Gal4 | F | 44955.11 | 0.56 |
| VGlut-F-200500 | 8793 | DvGlutMARCM-F003607_seg001 | VGlut-Gal4 | F | 75503.06 | 0.75 |
| VGlut-F-200176 | 10267 | DvGlutMARCM-F918_seg3 | VGlut-Gal4 | F | 68836.41 | 0.73 |
| VGlut-F-200395 | 11762 | DvGlutMARCM-F002333_seg001 | VGlut-Gal4 | F | 76400.37 | 0.71 |
| VGlut-F-500430 | 12098 | DvGlutMARCM-F002600_seg001 | VGlut-Gal4 | F | 78908.10 | 0.77 |
| VGlut-F-600350 | 12181 | DvGlutMARCM-F002659_seg001 | VGlut-Gal4 | F | 77597.12 | 0.76 |
| VGlut-F-000442 | 13078 | DvGlutMARCM-F2101_seg1 | VGlut-Gal4 | F | 76062.00 | 0.75 |
| VGlut-F-400502 | 13102 | DvGlutMARCM-F2127_seg1 | VGlut-Gal4 | F | 71431.34 | 0.63 |
| VGlut-F-200196 | 14380 | DvGlutMARCM-F1060_seg1 | VGlut-Gal4 | F | 75791.00 | 0.76 |
| VGlut-F-400361 | 14996 | DvGlutMARCM-F1337_seg1 | VGlut-Gal4 | F | 51843.38 | 0.53 |
| VGlut-F-500164 | 15100 | DvGlutMARCM-F1435_seg1 | VGlut-Gal4 | F | 76210.32 | 0.73 |
| VGlut-F-000385 | 15280 | DvGlutMARCM-F1606_seg2 | VGlut-Gal4 | F | 73150.75 | 0.74 |
| VGlut-F-400014 | 15696 | DvGlutMARCM-F111_seg1 | VGlut-Gal4 | F | 78073.53 | 0.76 |